Expanded geographic distribution for two Legionella pneumophila sequence types of clinical concern
- PMID: 39465969
- PMCID: PMC11580456
- DOI: 10.1128/msphere.00756-23
Expanded geographic distribution for two Legionella pneumophila sequence types of clinical concern
Abstract
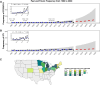
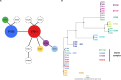
Legionella pneumophila serogroup 1 sequence types (ST) 213 and 222, a single-locus variant of ST213, were first detected in the early 1990s in the Midwest United States (U.S.) and the late 1990s in the Northeast U.S. and Canada. Since 1992, these STs have increasingly been implicated in community-acquired sporadic and outbreak-associated Legionnaires' disease (LD) cases. We were interested in understanding the change in LD frequency due to these STs and identifying genetic features that differentiate these STs from one another. For the geographic area examined here (Mountain West to Northeast) and over the study period (1992-2020), ST213/222-associated LD cases identified by the Centers for Disease Control and Prevention increased by 0.15 cases per year, with ST213/222-associated LD cases concentrated in four states: Michigan (26%), New York (18%), Minnesota (16%), and Ohio (10%). Additionally, between 2002 and 2021, ST222 caused at least five LD outbreaks in the U.S.; no known outbreaks due to ST213 occurred in the U.S. during this time. We compared the genomes of 230 ST213/222 isolates and found that the mean of the average nucleotide identity (ANI) within each ST was high (99.92% for ST222 and 99.92% for ST213), with a minimum between ST ANI of 99.50% and a maximum of 99.87%, indicating low genetic diversity within and between these STs. While genomic features were identified (e.g., plasmids and CRISPR-Cas systems), no association explained the increasing geographic distribution and prevalence of ST213 and ST222. Yet, we provide evidence of the expanded geographical distribution of ST213 and ST222 in the U.S.IMPORTANCESince the 1990s, cases of Legionnaires' disease (LD) attributed to a pair of closely related Legionella pneumophila variants, ST213 and ST222, have increased in the U.S. Furthermore, between 2002 and 2021, ST222 caused at least five outbreaks of LD in the U.S., while ST213 has not been linked to any U.S. outbreak. We wanted to understand how the rate of LD cases attributed to these variants has changed over time and compare the genetic features of the two variants. Between 1992 and 2020, we determined an increase of 0.15 LD cases ascribed to ST213/222 per year in the geographic region studied. Our research shows that these STs are spreading within the U.S., yet most of the cases occurred in four states: Michigan, New York, Minnesota, and Ohio. Additionally, we found little genetic diversity within and between these STs nor could specific genetic features explain their geographic spread.
Keywords: Legionnaires' disease; genomics; sequence types.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, Widmer A, Summersgill J, File T, Heath CM, Paterson DL, Chereshsky A. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community‐acquired legionellosis: an international collaborative survey. J Infect Dis 186:127–128. doi:10.1086/341087 - DOI - PubMed
-
- Gaia V, Fry NK, Afshar B, Lück PC, Meugnier H, Etienne J, Peduzzi R, Harrison TG. 2005. Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J Clin Microbiol 43:2047–2052. doi:10.1128/JCM.43.5.2047-2052.2005 - DOI - PMC - PubMed
-
- Ratzow S, Gaia V, Helbig JH, Fry NK, Lück PC. 2007. Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J Clin Microbiol 45:1965–1968. doi:10.1128/JCM.00261-07 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
