The T cell receptor β chain repertoire of tumor infiltrating lymphocytes improves neoantigen prediction and prioritization
- PMID: 39466298
- PMCID: PMC11517254
- DOI: 10.7554/eLife.94658
The T cell receptor β chain repertoire of tumor infiltrating lymphocytes improves neoantigen prediction and prioritization
Abstract
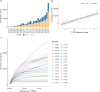
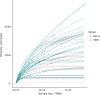
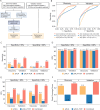
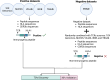
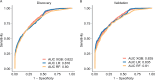
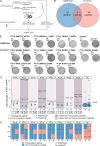
In the realm of cancer immunotherapy, the meticulous selection of neoantigens plays a fundamental role in enhancing personalized treatments. Traditionally, this selection process has heavily relied on predicting the binding of peptides to human leukocyte antigens (pHLA). Nevertheless, this approach often overlooks the dynamic interaction between tumor cells and the immune system. In response to this limitation, we have developed an innovative prediction algorithm rooted in machine learning, integrating T cell receptor β chain (TCRβ) profiling data from colorectal cancer (CRC) patients for a more precise neoantigen prioritization. TCRβ sequencing was conducted to profile the TCR repertoire of tumor-infiltrating lymphocytes (TILs) from 28 CRC patients. The data unveiled both intra-tumor and inter-patient heterogeneity in the TCRβ repertoires of CRC patients, likely resulting from the stochastic utilization of V and J segments in response to neoantigens. Our novel combined model integrates pHLA binding information with pHLA-TCR binding to prioritize neoantigens, resulting in heightened specificity and sensitivity compared to models using individual features alone. The efficacy of our proposed model was corroborated through ELISpot assays on long peptides, performed on four CRC patients. These assays demonstrated that neoantigen candidates prioritized by our combined model outperformed predictions made by the established tool NetMHCpan. This comprehensive assessment underscores the significance of integrating pHLA binding with pHLA-TCR binding analysis for more effective immunotherapeutic strategies.
Keywords: CRC; TCR sequencing; TCRseq; cancer biology; cancer immunotherapy; colorectal cancer; human; neoantigen; neoantigen prioritization; tumor variant calling.
© 2024, Pham et al.
Conflict of interest statement
TP, TN, BT, TD, ND, HP, TC, DL, HN, DT, TT, TP, ML, TV, MP, HG, HN, LT No competing interests declared
Figures





Update of
- doi: 10.1101/2023.11.16.567478
- doi: 10.7554/eLife.94658.1
- doi: 10.7554/eLife.94658.2
References
-
- 10x Genomics 2024a. CD8+ T cells of Healthy Donor 1. 10x Genomics. https://www.10xgenomics.com/resources/datasets/cd-8-plus-t-cells-of-heal...
-
- 10x Genomics 2024b. CD8+ T cells of Healthy Donor 2. 10x Genomics. https://www.10xgenomics.com/resources/datasets/cd-8-plus-t-cells-of-heal...
-
- 10x Genomics 2024c. CD8+ T cells of Healthy Donor 3. 10x Genomics. https://www.10xgenomics.com/resources/datasets/cd-8-plus-t-cells-of-heal...
-
- 10x Genomics 2024d. CD8+ T cells of Healthy Donor 4. 10x Genomics. https://www.10xgenomics.com/resources/datasets/cd-8-plus-t-cells-of-heal...
-
- Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The Eighth Edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA. 2017;67:93–99. doi: 10.3322/caac.21388. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

