Dexmedetomidine Ameliorates Myocardial Ischemia-Reperfusion Injury by Inhibiting MDH2 Lactylation via Regulating Metabolic Reprogramming
- PMID: 39467114
- PMCID: PMC11672254
- DOI: 10.1002/advs.202409499
Dexmedetomidine Ameliorates Myocardial Ischemia-Reperfusion Injury by Inhibiting MDH2 Lactylation via Regulating Metabolic Reprogramming
Abstract
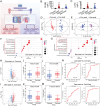
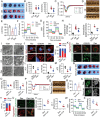
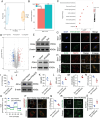
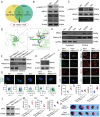
Myocardial ischemia-reperfusion injury (MIRI) significantly worsens the outcomes of patients with cardiovascular diseases. Dexmedetomidine (Dex) is recognized for its cardioprotective properties, but the related mechanisms, especially regarding metabolic reprogramming, have not been fully clarified. A total of 60 patients with heart valve disease are randomly assigned to Dex or control group. Blood samples are collected to analyze cardiac injury biomarkers and metabolomics. In vivo and vitro rat models of MIRI are utilized to assess the effects of Dex on cardiac function, lactate production, and mitochondrial function. It is found that postoperative CK-MB and cTNT levels are significantly lower in the Dex group. Metabolomics reveals that Dex regulates metabolic reprogramming and reduces lactate level. In Dex-treated rats, the myocardial infarction area is reduced, and myocardial contractility is improved. Dex inhibits glycolysis, reduces lactate, and improves mitochondrial function following MIRI. Lactylation proteomics identifies that Dex reduces the lactylation of Malate Dehydrogenase 2(MDH2), thus alleviating myocardial injury. Further studies reveal that MDH2 lactylation induces ferroptosis, leading to MIRI by impairing mitochondrial function. Mechanistic analyses reveal that Dex upregulates Nuclear Receptor Subfamily 3 Group C Member 1(NR3C1) phosphorylation, downregulates Pyruvate Dehydrogenase Kinase 4 (PDK4), and reduces lactate production and MDH2 lactylation. These findings provide new therapeutic targets and mechanisms for the treatment for MIRI.
Keywords: dexmedetomidine; ferroptosis; lactylation; metabolic reprogramming; myocardial ischemia‐reperfusion injury.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Role of Keap1-Nrf2/ARE signal transduction pathway in protection of dexmedetomidine preconditioning against myocardial ischemia/reperfusion injury.Biosci Rep. 2022 Sep 30;42(9):BSR20221306. doi: 10.1042/BSR20221306. Biosci Rep. 2022. PMID: 35959640 Free PMC article.
-
Dexmedetomidine Preconditioning Attenuates Myocardial Ischemia/Reperfusion Injury in Rats by Suppressing Mitophagy Via Activating Α2-Adrenergic Receptor.Arq Bras Cardiol. 2023 Oct;120(10):e20220750. doi: 10.36660/abc.20220750. Arq Bras Cardiol. 2023. PMID: 37909577 Free PMC article. English, Portuguese.
-
Dexmedetomidine Attenuates Myocardial Ischemia-Reperfusion Injury in Diabetes Mellitus by Inhibiting Endoplasmic Reticulum Stress.J Diabetes Res. 2019 Nov 30;2019:7869318. doi: 10.1155/2019/7869318. eCollection 2019. J Diabetes Res. 2019. PMID: 31886285 Free PMC article.
-
Dexmedetomidine inhibits pyroptosis by down-regulating miR-29b in myocardial ischemia reperfusion injury in rats.Int Immunopharmacol. 2020 Sep;86:106768. doi: 10.1016/j.intimp.2020.106768. Epub 2020 Jul 14. Int Immunopharmacol. 2020. PMID: 32679539
-
Lactylation: a promising therapeutic target in ischemia-reperfusion injury management.Cell Death Discov. 2025 Mar 13;11(1):100. doi: 10.1038/s41420-025-02381-4. Cell Death Discov. 2025. PMID: 40082399 Free PMC article. Review.
Cited by
-
Lactate-induced lactylation: from basic research to clinical perspectives.Front Pharmacol. 2025 Jun 13;16:1586973. doi: 10.3389/fphar.2025.1586973. eCollection 2025. Front Pharmacol. 2025. PMID: 40584617 Free PMC article. Review.
-
The interaction between ferroptosis and myocardial ischemia-reperfusion injury: molecular mechanisms and potential therapeutic targets.Eur J Med Res. 2025 Jul 21;30(1):643. doi: 10.1186/s40001-025-02851-6. Eur J Med Res. 2025. PMID: 40685346 Free PMC article. Review.
-
The Roles of Lactate and Lactylation in Diseases Related to Mitochondrial Dysfunction.Int J Mol Sci. 2025 Jul 24;26(15):7149. doi: 10.3390/ijms26157149. Int J Mol Sci. 2025. PMID: 40806283 Free PMC article. Review.
-
ACAT1-Mediated ME2 Acetylation Drives Chemoresistance in Ovarian Cancer by Linking Glutaminolysis to Lactate Production.Adv Sci (Weinh). 2025 Apr;12(14):e2416467. doi: 10.1002/advs.202416467. Epub 2025 Feb 14. Adv Sci (Weinh). 2025. PMID: 39951294 Free PMC article.
-
Lactate and lactylation in liver diseases: energy metabolism, inflammatory immunity and tumor microenvironment.Front Immunol. 2025 May 12;16:1581582. doi: 10.3389/fimmu.2025.1581582. eCollection 2025. Front Immunol. 2025. PMID: 40421024 Free PMC article. Review.
References
-
- Tsao C. W., Aday A. W., Almarzooq Z. I., Alonso A., Beaton A. Z., Bittencourt M. S., Boehme A. K., Buxton A. E., Carson A. P., Commodore‐Mensah Y., Elkind M. S. V., Evenson K. R., Eze‐Nliam C., Ferguson J. F., Generoso G., Ho J. E., Kalani R., Khan S. S., Kissela B. M., Knutson K. L., Levine D. A., Lewis T. T., Liu J., Loop M. S., Ma J., Mussolino M. E., Navaneethan S. D., Perak A. M., Poudel R., Rezk‐Hanna M., et al., Circulation 2022, 145, e153. - PubMed
-
- Bulluck H., Hausenloy D. J., Heart 2015, 101, 1067. - PubMed
-
- Heusch G., Nat. Rev. Cardiol. 2020, 17, 773. - PubMed
MeSH terms
Substances
Grants and funding
- 82300561/National Natural Science Foundation of China
- 82270523/National Natural Science Foundation of China
- 82305006/National Natural Science Foundation of China
- CSTB2023NSCQ-MSX0713/Natural Science Foundation of Chongqing
- YXQN202452/Chongqing Medical Young Talents Program
- YXQN202415/Chongqing Medical Young Talents Program
- High-Level Military Technological Innovation Talent Program
- CSTC2024YCJH-BGZXM0011/Chongqing Talent Program: Innovative Leading Talents
- CSTB2024NSCQ- LZX0079/Chongqing Natural Science Foundation Innovation and Development Joint Fund (Chongqing Education Commission)
- Sprint Program of Joint Medical Research of Chongqing Science and Chongqing Health Commission
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
