Perturbation-specific transcriptional mapping for unbiased target elucidation of antibiotics
- PMID: 39467118
- PMCID: PMC11551328
- DOI: 10.1073/pnas.2409747121
Perturbation-specific transcriptional mapping for unbiased target elucidation of antibiotics
Abstract
The rising prevalence of antibiotic resistance threatens human health. While more sophisticated strategies for antibiotic discovery are being developed, target elucidation of new chemical entities remains challenging. In the postgenomic era, expression profiling can play an important role in mechanism-of-action (MOA) prediction by reporting on the cellular response to perturbation. However, the broad application of transcriptomics has yet to fulfill its promise of transforming target elucidation due to challenges in identifying the most relevant, direct responses to target inhibition. We developed an unbiased strategy for MOA prediction, called perturbation-specific transcriptional mapping (PerSpecTM), in which large-throughput expression profiling of wild-type or hypomorphic mutants, depleted for essential targets, enables a computational strategy to address this challenge. We applied PerSpecTM to perform reference-based MOA prediction based on the principle that similar perturbations, whether chemical or genetic, will elicit similar transcriptional responses. Using this approach, we elucidated the MOAs of three molecules with activity against Pseudomonas aeruginosa by comparing their expression profiles to those of a reference set of antimicrobial compounds with known MOAs. We also show that transcriptional responses to small-molecule inhibition resemble those resulting from genetic depletion of essential targets by clustered regularly interspaced short palindromic repeats interference (CRISPRi) by PerSpecTM, demonstrating proof of concept that correlations between expression profiles of small-molecule and genetic perturbations can facilitate MOA prediction when no chemical entities exist to serve as a reference. Empowered by PerSpecTM, this work lays the foundation for an unbiased, readily scalable, systematic reference-based strategy for MOA elucidation that could transform antibiotic discovery efforts.
Keywords: RNAseq; antibiotics; gene expression; mechanism-of-action; transcriptomics.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

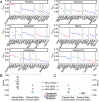
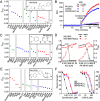
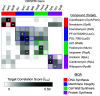
Update of
-
Perturbation-Specific Transcriptional Mapping for unbiased target elucidation of antibiotics.bioRxiv [Preprint]. 2024 Jun 7:2024.04.25.590978. doi: 10.1101/2024.04.25.590978. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2024 Nov 5;121(45):e2409747121. doi: 10.1073/pnas.2409747121. PMID: 38712067 Free PMC article. Updated. Preprint.
References
-
- Willyard C., The drug-resistant bacteria that pose the greatest health threats. Nature 543, 15 (2017). - PubMed
-
- Niu G., Li W., Next-Generation Drug Discovery to Combat Antimicrobial Resistance. Trends Biochem. Sci. 44, 961–972 (2019). - PubMed
-
- Lewis K., Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 12, 371–87 (2013). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

