Dysregulating mTORC1-4E-BP2 signaling in GABAergic interneurons impairs hippocampus-dependent learning and memory
- PMID: 39467641
- PMCID: PMC11606516
- DOI: 10.1101/lm.054018.124
Dysregulating mTORC1-4E-BP2 signaling in GABAergic interneurons impairs hippocampus-dependent learning and memory
Abstract
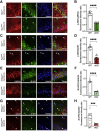
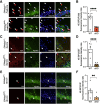
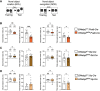
Memory formation is contingent on molecular and structural changes in neurons in response to learning stimuli-a process known as neuronal plasticity. The initiation step of mRNA translation is a gatekeeper of long-term memory by controlling the production of plasticity-related proteins in the brain. The mechanistic target of rapamycin complex 1 (mTORC1) controls mRNA translation, mainly through phosphorylation of the eukaryotic initiation factor 4E (eIF4E)-binding proteins (4E-BPs) and ribosomal protein S6 kinases (S6Ks). mTORC1 signaling decreases throughout brain development, starting from the early postnatal period. Here, we discovered that in mice, the age-dependent decrease in mTORC1 signaling occurs selectively in excitatory but not inhibitory neurons. Using a gene conditional knockout (cKO) strategy, we demonstrate that either up- or downregulating the mTORC1-4E-BP2 axis in GAD65 inhibitory interneurons, but not excitatory neurons, results in long-term object recognition and object location memory deficits. Our data indicate that the mTORC1 pathway in inhibitory but not excitatory neurons plays a key role in memory formation.
© 2024 Huang et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Amegandjin CA, Choudhury M, Jadhav V, Carriço JN, Quintal A, Berryer M, Snapyan M, Chattopadhyaya B, Saghatelyan A, Di Cristo G. 2021. Sensitive period for rescuing parvalbumin interneurons connectivity and social behavior deficits caused by TSC1 loss. Nat Commun 12: 3653. 10.1038/s41467-021-23939-7 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
