Comparative Analysis of Six Adeno-Associated Viral Vector Serotypes in Mouse Inferior Colliculus and Cerebellum
- PMID: 39467650
- PMCID: PMC11576142
- DOI: 10.1523/ENEURO.0391-24.2024
Comparative Analysis of Six Adeno-Associated Viral Vector Serotypes in Mouse Inferior Colliculus and Cerebellum
Abstract
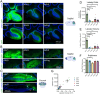
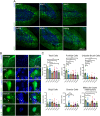
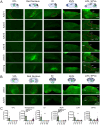
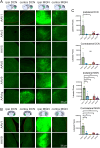
Adeno-associated viral vector (AAV) serotypes vary in how effectively they express genes across different cell types and brain regions. Here we report a systematic comparison of the AAV serotypes 1, 2, 5, 8, 9, and the directed evolution derived AAVrg, in the inferior colliculus (IC) and cerebellum. The AAVs were identical apart from their different serotypes, each having a synapsin promotor and expressing GFP (AAV-hSyn-GFP). Identical titers and volumes were injected into the IC and cerebellum of adult male and female mice, and brains were sectioned and imaged 2 weeks later. Transduction efficacy, anterograde labeling of axonal projections, and retrograde labeling of somata were characterized and compared across serotypes. Cell-type tropism was assessed by analyzing the morphology of the GFP-labeled neurons in the cerebellar cortex. In both the cerebellum and IC, AAV1 expressed GFP in more cells, labeled a larger volume, and produced significantly brighter labeling than all other serotypes, indicating superior transgene expression. AAV1 labeled more Purkinje cells, unipolar brush cells, and molecular layer interneurons than the other serotypes, while AAV2 labeled a greater number of granule cells. These results provide guidelines for the use of AAVs as gene delivery tools in these regions.
Keywords: adeno-associated virus; cerebellum; inferior colliculus.
Copyright © 2024 Witteveen and Balmer.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




Update of
-
Comparative analysis of six adeno-associated viral vector serotypes in mouse inferior colliculus and cerebellum.bioRxiv [Preprint]. 2024 Oct 21:2024.10.17.618966. doi: 10.1101/2024.10.17.618966. bioRxiv. 2024. Update in: eNeuro. 2024 Nov 4;11(11):ENEURO.0391-24.2024. doi: 10.1523/ENEURO.0391-24.2024. PMID: 39484622 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
