The Type 1 Diabetes T Cell Receptor and B Cell Receptor Repository in the AIRR Data Commons: a practical guide for access, use and contributions through the Type 1 Diabetes AIRR Consortium
- PMID: 39467874
- PMCID: PMC11663175
- DOI: 10.1007/s00125-024-06298-y
The Type 1 Diabetes T Cell Receptor and B Cell Receptor Repository in the AIRR Data Commons: a practical guide for access, use and contributions through the Type 1 Diabetes AIRR Consortium
Abstract
Human molecular genetics has brought incredible insights into the variants that confer risk for the development of tissue-specific autoimmune diseases, including type 1 diabetes. The hallmark cell-mediated immune destruction that is characteristic of type 1 diabetes is closely linked with risk conferred by the HLA class II gene locus, in combination with a broad array of additional candidate genes influencing islet-resident beta cells within the pancreas, as well as function, phenotype and trafficking of immune cells to tissues. In addition to the well-studied germline SNP variants, there are critical contributions conferred by T cell receptor (TCR) and B cell receptor (BCR) genes that undergo somatic recombination to yield the Adaptive Immune Receptor Repertoire (AIRR) responsible for autoimmunity in type 1 diabetes. We therefore created the T1D TCR/BCR Repository (The Type 1 Diabetes T Cell Receptor and B Cell Receptor Repository) to study these highly variable and dynamic gene rearrangements. In addition to processed TCR and BCR sequences, the T1D TCR/BCR Repository includes detailed metadata (e.g. participant demographics, disease-associated parameters and tissue type). We introduce the Type 1 Diabetes AIRR Consortium goals and outline methods to use and deposit data to this comprehensive repository. Our ultimate goal is to facilitate research community access to rich, carefully annotated immune AIRR datasets to enable new scientific inquiry and insight into the natural history and pathogenesis of type 1 diabetes.
Keywords: AIRR; AIRR Data Commons; Autoantibodies; B cell receptors; FAIR data; Next-generation sequencing; Single-cell RNA-seq; T cell receptors; Type 1 diabetes.
© 2024. The Author(s).
Conflict of interest statement
Acknowledgements: The authors would like to acknowledge the contributions of the(sugar)science, a type 1 diabetes research advocacy organisation, for initiating, organising and hosting the consortium activities. All screen grabs of the iReceptor Gateway are provided with the permission of the iReceptor project. Type 1 Diabetes AIRR Consortium members: Erin Baschal [Barbara Davis Center for Diabetes, University of Colorado School of Medicine, Aurora, CO 80045, USA], Karen Cerosaletti [Center for Translational Immunology, Benaroya Research Institute at Virginia Mason, Seattle, WA], Lorissa Corrie [iReceptor Genomic Services, Summerland, BC, Canada], Iria Gomez-Tourino [Centre for Research in Molecular Medicine and Chronic Diseases (CiMUS), University of Santiago de Compostela, Santiago de Compostela, Spain], Lauren Higdon [Diabetes Center, University of California San Francisco, San Francisco, CA, USA], Sally C. Kent [Diabetes Center of Excellence, Department of Medicine, University of Massachusetts Medical Chan School, Worcester, MA, USA], Peter Linsley [Benaroya Research Institute, Seattle, WA 98101, USA], Maki Nakayama [Barbara Davis Center for Diabetes, University of Colorado School of Medicine, Aurora, CO 80045, USA], Kira Neller [iReceptor Genomic Services, Summerland, BC, Canada], William E. Ruff [Repertoire Immune Medicines, Cambridge, MA 02139, USA], Luc Teyton [Department of Immunology and Microbiology, Scripps Research, La Jolla, CA 92037, USA] Data availability: All data in Table 2 are available from https://gateway.ireceptor.org/login Data from Fig. 7 are available at https://app.litmaps.com/shared/12ead8ba-35d9-4232-91a0-26da9e3c3f59 Funding: SJH is funded by the Diabetes Research and Wellness Foundation Professor David Matthews Non-Clinical Research Fellowship, The Leona M. and Harry B. Helmsley Charitable Trust by grant #2101-04969, and this work was supported on behalf of the “Steve Morgan Foundation Type 1 Diabetes Grand Challenge” by Breakthrough T1D UK, formerly JDRF and SMF (grant numbers 2-SRA-2024-1474-M-N and 2-SRA-2024-1473-M-N). This work was also funded by NIH grants R01 DK131070 (RHB), DK108868 (AWM), DK032083 (AWM), DK099317 (AWM), U24 AI177622 (FB, BC) and U01-DK112217 (ETLP). TMB is funded by NIH P01 AI042288 and AWM and TMB by The Leona M. and Harry B. Helmsley Charitable Trust by grant #2301-06562. Authors’ relationships and activities: TMB has consulted for Repertoire Immune Medicines and both TMB and AWM have received in-kind sequencing support by Adaptive Biotech. BC and FB are both researchers on the iReceptor team at Simon Fraser University and are partners in iReceptor Genomic Services. The other authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work. Contribution statement: SJH, RHB, ALP, ETLP, MW, FB, AWM and TMB were responsible for conceptualisation and design of the work. BC was responsible for data curation: MW provided project administration for the Type 1 Diabetes AIRR Consortium. MW, SJH, RHB and BC wrote the original draft. All authors reviewed and edited the original draft and approved the final manuscript for publication. TMB and AWM are guarantors of this work
Figures
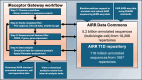


References
-
- Emerson RO, DeWitt WS, Vignali M et al (2017) Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet 49(5):659–665. 10.1038/ng.3822 - PubMed
MeSH terms
Substances
Grants and funding
- P01 AI042288/GF/NIH HHS/United States
- P01 AI042288/AI/NIAID NIH HHS/United States
- Professor David Matthews Non-Clinical Research Fel/Diabetes Research and Wellness Foundation
- DK108868/GF/NIH HHS/United States
- DK131070 DK032083 DK099317/GF/NIH HHS/United States
- #2101-04969 #2301-06562/Leona M. and Harry B. Helmsley Charitable Trust
- U01-DK112217/GF/NIH HHS/United States
- U24 AI177622/GF/NIH HHS/United States
- 2-SRA-2024-1473-M-N/Steve Morgan Grand Chellenge/JDRF
- R01 DK032083/DK/NIDDK NIH HHS/United States
- R01 DK108868/DK/NIDDK NIH HHS/United States
- R01 DK099317/DK/NIDDK NIH HHS/United States
- 2-SRA-2024-1474-M-N/Steve Morgan Grand Chellenge/JDRF
- R01 DK131070/DK/NIDDK NIH HHS/United States
- U24 AI177622/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

