Phospho-signaling couples polar asymmetry and proteolysis within a membraneless microdomain in Caulobacter crescentus
- PMID: 39468040
- PMCID: PMC11519897
- DOI: 10.1038/s41467-024-53395-y
Phospho-signaling couples polar asymmetry and proteolysis within a membraneless microdomain in Caulobacter crescentus
Abstract
Asymmetric cell division in bacteria is achieved through cell polarization, where regulatory proteins are directed to specific cell poles. In Caulobacter crescentus, both poles contain a membraneless microdomain, established by the polar assembly hub PopZ, through most of the cell cycle, yet many PopZ clients are unipolar and transiently localized. We find that PopZ's interaction with the response regulator CpdR is controlled by phosphorylation, via the histidine kinase CckA. Phosphorylated CpdR does not interact with PopZ and is not localized to cell poles. At poles where CckA acts as a phosphatase, dephosphorylated CpdR binds directly with PopZ and subsequently recruits ClpX, substrates, and other members of a protease complex to the cell pole. We also find that co-recruitment of protease components and substrates to polar microdomains enhances their coordinated activity. This study connects phospho-signaling with polar assembly and the activity of a protease that triggers cell cycle progression and cell differentiation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

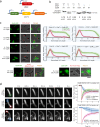
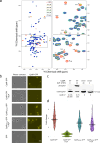
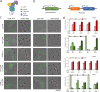


Update of
-
Phospho-signaling couples polar asymmetry and proteolysis within a membraneless microdomain in C. crescentus.bioRxiv [Preprint]. 2023 Aug 20:2023.08.19.553945. doi: 10.1101/2023.08.19.553945. bioRxiv. 2023. Update in: Nat Commun. 2024 Oct 28;15(1):9282. doi: 10.1038/s41467-024-53395-y. PMID: 37645878 Free PMC article. Updated. Preprint.
References
-
- Van Teeseling, M. C. F. & Thanbichler, M. Generating asymmetry in a changing environment: Cell cycle regulation in dimorphic alphaproteobacteria. Biol. Chem. 401, 1349–1363 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 GM113131/GM/NIGMS NIH HHS/United States
- DBI-1828319/National Science Foundation (NSF)
- R01GM118792/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- P20 GM103432/GM/NIGMS NIH HHS/United States
- 2P20GM103432/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
LinkOut - more resources
Full Text Sources

