Conserved autism-associated genes tune social feeding behavior in C. elegans
- PMID: 39468047
- PMCID: PMC11519495
- DOI: 10.1038/s41467-024-53590-x
Conserved autism-associated genes tune social feeding behavior in C. elegans
Abstract
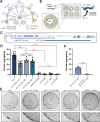
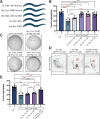
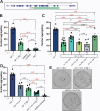
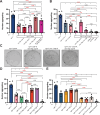
Animal foraging is an essential and evolutionarily conserved behavior that occurs in social and solitary contexts, but the underlying molecular pathways are not well defined. We discover that conserved autism-associated genes (NRXN1(nrx-1), NLGN3(nlg-1), GRIA1,2,3(glr-1), GRIA2(glr-2), and GLRA2,GABRA3(avr-15)) regulate aggregate feeding in C. elegans, a simple social behavior. NRX-1 functions in chemosensory neurons (ADL and ASH) independently of its postsynaptic partner NLG-1 to regulate social feeding. Glutamate from these neurons is also crucial for aggregate feeding, acting independently of NRX-1 and NLG-1. Compared to solitary counterparts, social animals show faster presynaptic release and more presynaptic release sites in ASH neurons, with only the latter requiring nrx-1. Disruption of these distinct signaling components additively converts behavior from social to solitary. Collectively, we find that aggregate feeding is tuned by conserved autism-associated genes through complementary synaptic mechanisms, revealing molecular principles driving social feeding.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures
Update of
-
Conserved autism-associated genes tune social feeding behavior in C. elegans.bioRxiv [Preprint]. 2023 Dec 6:2023.12.05.570116. doi: 10.1101/2023.12.05.570116. bioRxiv. 2023. Update in: Nat Commun. 2024 Oct 28;15(1):9301. doi: 10.1038/s41467-024-53590-x. PMID: 38106124 Free PMC article. Updated. Preprint.
References
-
- Gordon D. M. Ant Encounters: Interaction Networks and Colony Behavior. In: Ant Encounters. Princeton (University Press, 2010).
-
- Azorsa, F., Muscedere, M. L. & Traniello, J. F. A. Socioecology and evolutionary neurobiology of predatory ants. Front. Ecol. Evol. 9, 10.3389/fevo.2021.804200 (2022).
Publication types
MeSH terms
Substances
Grants and funding
- R56 MH096881/MH/NIMH NIH HHS/United States
- 1R35GM146782/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R35 GM146782/GM/NIGMS NIH HHS/United States
- 1R01MH096881/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- P40 OD010440/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

