Clinical genome sequencing in patients with suspected rare genetic disease in Peru
- PMID: 39468051
- PMCID: PMC11519459
- DOI: 10.1038/s41525-024-00434-8
Clinical genome sequencing in patients with suspected rare genetic disease in Peru
Abstract
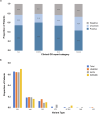
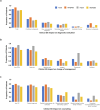
There is limited access to molecular genetic testing in most low- and middle-income countries. The iHope program provides clinical genome sequencing (cGS) to underserved individuals with signs or symptoms of rare genetic diseases and limited or no access to molecular genetic testing. Here we describe the performance and impact of cGS in 247 patients from three clinics in Peru. Although most patients had at least one genetic test prior to cGS (70.9%), the most frequent was karyotyping (53.4%). The diagnostic yield of cGS was 54.3%, with candidate variants reported in an additional 22.3% of patients. Clinical GS results impacted clinician diagnostic evaluation in 85.0% and genetic counseling in 72.1% of cases. Changes in management were reported in 71.3%, inclusive of referrals (64.7%), therapeutics (26.3%), laboratory or physiological testing (25.5%), imaging (19%), and palliative care (17.4%), suggesting that increased availability of genomic testing in Peru would enable improved patient management.
© 2024. The Author(s).
Conflict of interest statement
Ryan J. Taft, Erin Thorpe, Subramanian S. Ajay, James Avecilla, Krista Bluske, Carolyn M. Brown, Amanda Buchanan, Brendan Burns, Nicole Burns, Anjana Chandrasekhar, Amanda Clause, Katie Golden-Grant, R. Tanner Hagelstrom, Rueben Hejja, Basil Juan, Alka Malhotra, Philip Medrano, Becky Milewski, Felipe Mullen, Viswateja Nelakuditi, Vani Rajan, Revathi Rajkumar, Samin Sajan, Zinayida Schlachetzki, Sarah Schmidt, Julie Taylor, and Brittany Thomas, Evgenii Chekalin, Max Arseneault, Maren Bennett, Aditi Chawla, Alison J. Coffey, Akanchha Kesari, Denise L. Perry, Ajay Ramakrishnan Sylwia Urbaniak, Andrew Warren were employees of and stockholders in Illumina, Inc. during this study.
Figures



Similar articles
-
The impact of clinical genome sequencing in a global population with suspected rare genetic disease.Am J Hum Genet. 2024 Jul 11;111(7):1271-1281. doi: 10.1016/j.ajhg.2024.05.006. Epub 2024 Jun 5. Am J Hum Genet. 2024. PMID: 38843839 Free PMC article.
-
The implementation of genome sequencing in rare genetic diseases diagnosis: a pilot study from the Hong Kong genome project.Lancet Reg Health West Pac. 2025 Jan 28;55:101473. doi: 10.1016/j.lanwpc.2025.101473. eCollection 2025 Feb. Lancet Reg Health West Pac. 2025. PMID: 39944418 Free PMC article.
-
Randomized prospective evaluation of genome sequencing versus standard-of-care as a first molecular diagnostic test.Genet Med. 2021 Sep;23(9):1689-1696. doi: 10.1038/s41436-021-01193-y. Epub 2021 May 11. Genet Med. 2021. PMID: 33976420 Free PMC article. Clinical Trial.
-
Case for genome sequencing in infants and children with rare, undiagnosed or genetic diseases.J Med Genet. 2019 Dec;56(12):783-791. doi: 10.1136/jmedgenet-2019-106111. Epub 2019 Apr 25. J Med Genet. 2019. PMID: 31023718 Free PMC article. Review.
-
Genetic Counseling and Genome Sequencing in Pediatric Rare Disease.Cold Spring Harb Perspect Med. 2020 Mar 2;10(3):a036632. doi: 10.1101/cshperspect.a036632. Cold Spring Harb Perspect Med. 2020. PMID: 31501267 Free PMC article. Review.
References
-
- Instituto Nacional de Estadistica E. Informatica. Censos Nacionales 2017: XII de Poblacion, VII Vivienda y III de Comunidades Indigenas. https://www.inei.gob.pe/media/MenuRecursivo/publicaciones_digitales/Est/... (2017).
-
- Azam, M. Governance and economic growth: evidence from 14 Latin America and Caribbean Countries. J. Knowl. Econ.13, 1470–1495 (2022).
-
- Varona-Castillo, L. & Gonzales-Castillo, J. R. Crecimiento económico y distribución del ingreso en Perú. Probl. Desarro. Rev. Latinoam. Econ. 52, 79–107 (2021).
-
- Cambra-Fierro, J. J., Fuentes-Blasco, M., Huerta-Álvarez, R. & Olavarría-Jaraba, A. Destination recovery during COVID-19 in an emerging economy: Insights from Perú. Eur. Res. Manag. Bus. Econ.28, 100188 (2022).
LinkOut - more resources
Full Text Sources

