Regeneration-specific promoter switching facilitates Mest expression in the mouse digit tip to modulate neutrophil response
- PMID: 39468052
- PMCID: PMC11519450
- DOI: 10.1038/s41536-024-00376-w
Regeneration-specific promoter switching facilitates Mest expression in the mouse digit tip to modulate neutrophil response
Abstract
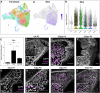
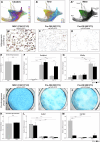
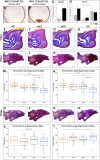
The mouse digit tip regenerates following amputation, a process mediated by a cellularly heterogeneous blastema. We previously found the gene Mest to be highly expressed in mesenchymal cells of the blastema and a strong candidate pro-regenerative gene. We now show Mest digit expression is regeneration-specific and not upregulated in post-amputation fibrosing proximal digits. Mest homozygous knockout mice exhibit delayed bone regeneration though no phenotype is found in paternal knockout mice, inconsistent with the defined maternal genomic imprinting of Mest. We demonstrate that promoter switching, not loss of imprinting, regulates biallelic Mest expression in the blastema and does not occur during embryogenesis, indicating a regeneration-specific mechanism. Requirement for Mest expression is tied to modulating neutrophil response, as revealed by scRNAseq and FACS comparing wildtype and knockout blastemas. Collectively, the imprinted gene Mest is required for proper digit tip regeneration and its blastema expression is facilitated by promoter switching for biallelic expression.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
Regeneration-specific promoter switching facilitates Mest expression in the mouse digit tip to modulate neutrophil response.bioRxiv [Preprint]. 2024 Jun 15:2024.06.12.598713. doi: 10.1101/2024.06.12.598713. bioRxiv. 2024. Update in: NPJ Regen Med. 2024 Oct 28;9(1):32. doi: 10.1038/s41536-024-00376-w. PMID: 38915675 Free PMC article. Updated. Preprint.
References
Grants and funding
- F31 AR082220/AR/NIAMS NIH HHS/United States
- R01 HD109200/HD/NICHD NIH HHS/United States
- T32 AR055885/AR/NIAMS NIH HHS/United States
- DP-0205-2201/Harvard University | Harvard Stem Cell Institute (HSCI)
- F31AR082220/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
LinkOut - more resources
Full Text Sources

