Light inducible gene expression system for Streptomyces
- PMID: 39468183
- PMCID: PMC11519972
- DOI: 10.1038/s41598-024-76860-6
Light inducible gene expression system for Streptomyces
Abstract
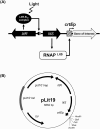
The LitR/CarH family comprises adenosyl B12-based photosensory transcriptional regulators that control light-inducible carotenoid production in nonphototrophic bacteria. In this study, we established a blue-green light-inducible hyperexpression system using LitR and its partner ECF-type sigma factor LitS in streptomycin-producing Streptomyces griseus NBRC 13350. The constructed multiple-copy number plasmid, pLit19, carried five genetic elements: pIJ101rep, the thiostrepton resistance gene, litR, litS, and σLitS-recognized light-inducible crtE promoter. Streptomyces griseus transformants harboring pLit19 exhibited a light-dependent hyper-production of intracellular reporter enzymes including catechol-2,3-dioxygenase and β-glucuronidase, extracellular secreted enzymes including laccase and transglutaminase, and secondary metabolites including melanin, flaviolin, and indigoidine. Cephamycin-producing Streptomyces sp. NBRC 13304, carrying an entire actinorhodin gene cluster, exhibited light-dependent actinorhodin production after the introduction of the pLit19 shuttle-type plasmid with the pathway-specific activator actII-ORF4. Insertion of sti fragment derived from Streptomyces phaeochromogenes pJV1 plasmid into pLit19 increased its light sensitivity, allowing gene expression under weak light irradiation. The two constructed Escherichia coli-Streptomyces shuttle-type pLit19 plasmids were found to have abilities similar to those of pLit19. We successfully established an optogenetically controlled hyperproduction system for S. griseus NBRC 13350 and Streptomyces sp. NBRC 13304.
Keywords: Streptomyces; Light-inducible; LitR; Optogenetics; Recombinant protein.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Beg, Q. K., Kapoor, M., Mahajan, L. & Hoondal, G. S. Microbial xylanases and their industrial applications: a review. Appl. Microbiol. Biotechnol.56, 326–38 (2001). - PubMed
-
- Fuchsbauer, H.-L. Approaching transglutaminase from Streptomyces bacteria over three decades. FEBS J.289, 4680–4703 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

