A novel risk score model of lactate metabolism for predicting outcomes and immune signatures in acute myeloid leukemia
- PMID: 39468216
- PMCID: PMC11519446
- DOI: 10.1038/s41598-024-76919-4
A novel risk score model of lactate metabolism for predicting outcomes and immune signatures in acute myeloid leukemia
Abstract
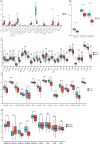
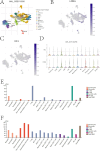
Acute myeloid leukemia (AML) is a malignant tumor with high recurrence and refractory rates and low survival rates. Increased glycolysis is characteristic of metabolism in AML blast cells and is also associated with chemotherapy resistance. The purpose of this study was to use gene expression and clinical information from The Cancer Genome Atlas (TCGA) database to identify subtypes of AML associated with lactate metabolism. Two different subtypes linked to lactate metabolism, each with specific immunological features and consequences for prognosis, were identified in this study. Using the TCGA and International Cancer Genome Consortium (GEO) cohorts, a prognostic model composed of genes (LMNA, RETN and HK1) for the prognostic value of the lactate metabolism-related risk score prognostic model was created and validated, suggesting possible therapeutic uses. Additionally, the diagnostic value of the prognostic model genes was explored. LMNA and HK1 were ultimately identified as hub genes, and their roles in AML were determined through immune infiltration, GeneMANIA, GSEA, methylation analysis and single-cell analysis. LMNA was upregulated in AML associating with a poor prognosis while HK1 was downregulated in AML associating with a favorable prognosis. The findings underscore the noteworthy impact of genes linked to lactate metabolism in AML and illustrate the possible therapeutic usefulness of the lactate metabolism-related risk score and the hub lactate metabolism-related genes in guiding AML patients' treatment choices.
Keywords: Acute myeloid leukemia; Immune signature; Lactate metabolism-associated gene; OS; Risk model.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Pollyea, D. A. et al. Acute myeloid leukemia, Version 3.2023, NCCN Clinical Practice guidelines in Oncology. J. Natl. Compr. Cancer Netw. JNCCN 21, 503–513. 10.6004/jnccn.2023.0025 (2023). - PubMed
-
- Herst, P. M., Howman, R. A., Neeson, P. J., Berridge, M. V. & Ritchie, D. S. The level of glycolytic metabolism in acute myeloid leukemia blasts at diagnosis is prognostic for clinical outcome. J. Leukoc. Biol. 89, 51–55. 10.1189/jlb.0710417 (2011). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

