Activation of the general stress response sigma factor SigB prevents competence development in Bacillus subtilis
- PMID: 39470193
- PMCID: PMC11633097
- DOI: 10.1128/mbio.02274-24
Activation of the general stress response sigma factor SigB prevents competence development in Bacillus subtilis
Abstract
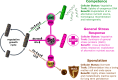
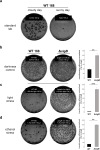
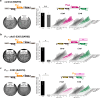
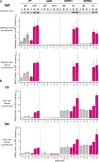
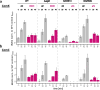
Seemingly simple bacteria mount intricate adaptive responses when exposed to physical stress or nutrient limitation, and the activation of these responses is governed by complex signal transduction networks. Upon entry into the stationary growth phase, the soil bacterium Bacillus subtilis may develop natural competence, form biofilms or stress-resistant cells, or ultimately trigger a cellular differentiation program leading to spore formation. Master regulators, such as Spo0A, ComK, SinR, and SigB, constantly monitor the bacterium's environment and then determine appropriate adaptive responses. Here, we show that exposure of B. subtilis to visible light and other stresses triggers a general stress response-dependent block in competence development. SigB serves as an "emergency system" to silence inappropriate expression of an alternative developmental program in the face of unfavorable conditions. In particular, we document a stress-dependent molecular mechanism that prevents accumulation of the central competence regulator ComK via expression of a SigB-driven antisense RNA (as-comK, S365) which is part of a noncontiguous operon.
Importance: Bacillus subtilis exhibits a large number of different specific and general adaptation reactions, which need to be well balanced to sustain survival under largely unfavorable conditions. Under specific conditions, natural competence develops, which enables B. subtilis to actively take up exogenous DNA to integrate it into its own genome. In contrast to this specific adaptation, the general stress response is induced by a variety of exogenous stress and starvation stimuli, providing comprehensive protection and enabling survival of vegetative B. subtilis cells. In the present work, we reveal the molecular basis for the interconnection of these two important responses in the regulatory network. We describe that the master regulator of the general stress response SigB is activated by physiological stress stimuli, including daylight and ethanol stress, leading to the inactivation of the competence master regulator ComK by transcriptional anti-sense regulation, showing a strict hierarchy of adaptational responses under severe stress.
Keywords: ComK; ComK anti-sense RNA; SigB; Spo0A; as-comK (feature S365); competence development; general stress response; sporulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

