Structures of Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus virions reveal species-specific tegument and envelope features
- PMID: 39470208
- PMCID: PMC11575322
- DOI: 10.1128/jvi.01194-24
Structures of Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus virions reveal species-specific tegument and envelope features
Abstract
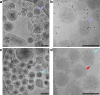
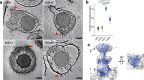
Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) are classified into the gammaherpesvirus subfamily of Herpesviridae, which stands out from its alpha- and betaherpesvirus relatives due to the tumorigenicity of its members. Although structures of human alpha- and betaherpesviruses by cryogenic electron tomography (cryoET) have been reported, reconstructions of intact human gammaherpesvirus virions remain elusive. Here, we structurally characterize extracellular virions of EBV and KSHV by deep learning-enhanced cryoET, resolving both previously known monomorphic capsid structures and previously unknown pleomorphic features beyond the capsid. Through subtomogram averaging and subsequent tomogram-guided sub-particle reconstruction, we determined the orientation of KSHV nucleocapsids from mature virions with respect to the portal to provide spatial context for the tegument within the virion. Both EBV and KSHV have an eccentric capsid position and polarized distribution of tegument. Tegument species span from the capsid to the envelope and may serve as scaffolds for tegumentation and envelopment. The envelopes of EBV and KSHV are less densely populated with glycoproteins than those of herpes simplex virus 1 (HSV-1) and human cytomegalovirus (HCMV), representative members of alpha- and betaherpesviruses, respectively. Also, we observed fusion protein gB trimers exist within triplet arrangements in addition to standalone complexes, which is relevant to understanding dynamic processes such as fusion pore formation. Taken together, this study reveals nuanced yet important differences in the tegument and envelope architectures among human herpesviruses and provides insights into their varied cell tropism and infection.
Importance: Discovered in 1964, Epstein-Barr virus (EBV) is the first identified human oncogenic virus and the founding member of the gammaherpesvirus subfamily. In 1994, another cancer-causing virus was discovered in lesions of AIDS patients and later named Kaposi's sarcoma-associated herpesvirus (KSHV), the second human gammaherpesvirus. Despite the historical importance of EBV and KSHV, technical difficulties with isolating large quantities of these viruses and the pleiomorphic nature of their envelope and tegument layers have limited structural characterization of their virions. In this study, we employed the latest technologies in cryogenic electron microscopy (cryoEM) and tomography (cryoET) supplemented with an artificial intelligence-powered data processing software package to reconstruct 3D structures of the EBV and KSHV virions. We uncovered unique properties of the envelope glycoproteins and tegument layers of both EBV and KSHV. Comparison of these features with their non-tumorigenic counterparts provides insights into their relevance during infection.
Keywords: Epstein-Barr virus; Kaposi's sarcoma-associated herpesvirus; electron microscopy; glycoproteins; herpesviruses; human herpesviruses; virion structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Update of
-
Structures of Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus virions reveal species-specific tegument and envelope features.bioRxiv [Preprint]. 2024 Jul 9:2024.07.09.602672. doi: 10.1101/2024.07.09.602672. bioRxiv. 2024. Update in: J Virol. 2024 Nov 19;98(11):e0119424. doi: 10.1128/jvi.01194-24. PMID: 39026862 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
- R21AI175798/HHS | National Institutes of Health (NIH)
- R21 AI175798/AI/NIAID NIH HHS/United States
- R01DE027901/HHS | National Institutes of Health (NIH)
- R01DE025567/HHS | National Institutes of Health (NIH)
- IRG-15-173-21/American Cancer Society (ACS)
- P30 CA060553/CA/NCI NIH HHS/United States
- U24 GM116792/GM/NIGMS NIH HHS/United States
- R01 DE025567/DE/NIDCR NIH HHS/United States
- T32 AI060567/AI/NIAID NIH HHS/United States
- S10 RR023057/RR/NCRR NIH HHS/United States
- R01 DE027901/DE/NIDCR NIH HHS/United States
- S10 OD018111/OD/NIH HHS/United States
- 5T32AI060567/HHS | National Institutes of Health (NIH)
LinkOut - more resources
Full Text Sources

