Biofilm-mediated antibiotic tolerance in Staphylococcus aureus from spinal cord stimulation device-related infections
- PMID: 39470274
- PMCID: PMC11619394
- DOI: 10.1128/spectrum.01683-24
Biofilm-mediated antibiotic tolerance in Staphylococcus aureus from spinal cord stimulation device-related infections
Abstract
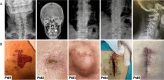
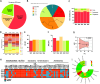
Staphylococcus aureus is a predominant cause of infections in individuals with spinal cord stimulation (SCS) devices. Biofilm formation complicates these infections, commonly requiring both surgical and antibiotic treatments. This study explored the biofilm matrix composition and antimicrobial susceptibility of planktonic and biofilm-growing S. aureus isolates from individuals with SCS-related infections. Whole-genome sequencing (WGS) examined genotypes, virulome, resistome, and the pan-genome structure. The study also analyzed biofilm matrix composition, early surface adhesion, hemolytic activity, and antibiotic-susceptibility testing. WGS revealed genetic diversity among isolates. One isolate, though oxacillin susceptible, contained the mecA gene. The median number of virulence factor genes per isolate was 58. All isolates harbored the biofilm-related icaA/D genes. When assessing phenotypic characteristics, all strains demonstrated the ability to form biofilms in vitro. The antimicrobial susceptibility profile indicated that oxacillin, rifampin, and teicoplanin showed the highest efficacy against S. aureus biofilm. Conversely, high biofilm tolerance was observed for vancomycin, trimethoprim/sulfamethoxazole, and levofloxacin. These findings suggest that S. aureus isolates are highly virulent and produce robust biofilms. In cases of suspected biofilm infections caused by S. aureus, vancomycin should not be the primary choice due to its low activity against biofilm. Instead, oxacillin, rifampin, and teicoplanin appear to be more effective options to manage SCS infections.IMPORTANCESCS devices are increasingly used to manage chronic pain, but infections associated with these devices, particularly those caused by Staphylococcus aureus, present significant clinical challenges. These infections are often complicated by biofilm formation, which protects bacteria from immune responses and antibiotic treatments, making them difficult to eradicate. Understanding the genetic diversity, virulence, and biofilm characteristics of S. aureus isolates from SCS infections is critical to improving treatment strategies. Our study highlights the need to reconsider commonly used antibiotics like vancomycin, which shows reduced activity against biofilm-growing cells. Identifying more effective alternatives, such as oxacillin, rifampin, and teicoplanin, provides valuable insight for clinicians when managing biofilm-related S. aureus infections in patients with SCS implants. This research contributes to the growing evidence that biofilm formation is crucial in treating device-related infections, emphasizing the importance of tailoring antimicrobial strategies to the biofilm phenotype.
Keywords: Staphylococcus aureus; biofilm; oxacillin; spinal cord stimulation; vancomycin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

