Real-time plasmid transmission detection pipeline
- PMID: 39470278
- PMCID: PMC11619237
- DOI: 10.1128/spectrum.02100-24
Real-time plasmid transmission detection pipeline
Abstract
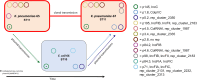
The spread of antimicrobial resistance among bacteria by horizontal plasmid transmissions poses a major challenge for clinical microbiology. Here, we evaluate a new real-time plasmid transmission detection pipeline implemented in the SeqSphere+ (Ridom GmbH, Münster, Germany) software. Within the pipeline, a local Mash plasmid database is created, and Mash searches with a distance threshold of 0.001 are used to trigger plasmid transmission early warning alerts (EWAs). Clonal transmissions are detected using core-genome multi-locus sequence typing allelic differences. The tools MOB-suite, NCBI AMRFinderPlus, CGE MobileElementFinder, pyGenomeViz, and MUMmer, integrated in SeqSphere+, are used to characterize plasmids and for visual pairwise plasmid comparisons, respectively. We evaluated the pipeline using published hybrid assemblies (Oxford Nanopore Technology/Illumina) of a surveillance and outbreak data set with plasmid transmissions. To emulate prospective usage, samples were imported in chronological order of sampling date. Different combinations of the user-adjustable parameters sketch size (1,000 vs 10,000) and plasmid size correction were tested, and discrepancies between resulting clusters were analyzed with Quast. When using a sketch size of 1,000 with size correction turned on, the SeqSphere+ pipeline agreed with the published data and produced the same clonal and carbapenemase-carrying plasmid clusters. EWAs were in the correct chronological order. In summary, the developed pipeline presented here is suitable for integration into clinical microbiology settings with limited bioinformatics knowledge due to its automated analyses and alert system, which are combined with the GUI-based SeqSphere+ platform. Thus, with its integrated sample database, (near) real-time plasmid transmission detection is within reach in bacterial routine-diagnostic settings when long-read sequencing is employed.
Importance: Plasmid-mediated spread of antimicrobial resistance is a major challenge for clinical microbiology, and monitoring of potential plasmid transmissions is essential to combat further dissemination. Whole-genome sequencing is often used to surveil nosocomial transmissions but usually limited to the detection of clonal transmissions (based on chromosomal markers). Recent advances in long-read sequencing technologies enable full reconstruction of plasmids and the detection of very similar plasmids, but so far, easy-to-use bioinformatic tools for this purpose have been missing. Here, we present an evaluation of an innovative real-time plasmid transmission detection pipeline. It is integrated into the GUI-based SeqSphere+ software, which already offers core-genome multi-locus sequence typing-based pathogen outbreak detection. It requires very limited bioinformatics knowledge, and its database, automated analyses, and alert system make it well suited for prospective clinical application.
Keywords: antimicrobial resistance; horizontal gene transfer; long-read sequencing; plasmid; transmission; whole-genome sequencing.
Conflict of interest statement
N.S., J.R., and T.W. are (part-time) employees of Ridom GmbH. J.R., T.W., and D.H. are shareholders of Ridom GmbH. A.M. declares no conflict of interest.
Figures


References
-
- Sheppard AE, Stoesser N, Wilson DJ, Sebra R, Kasarskis A, Anson LW, Giess A, Pankhurst LJ, Vaughan A, Grim CJ, Cox HL, Yeh AJ, Sifri CD, Walker AS, Peto TE, Crook DW, Mathers AJ, Modernising Medical Microbiology (MMM) Informatics Group . 2016. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob Agents Chemother 60:3767–3778. doi:10.1128/AAC.00464-16 - DOI - PMC - PubMed
-
- Orlek A, Stoesser N, Anjum MF, Doumith M, Ellington MJ, Peto T, Crook D, Woodford N, Walker AS, Phan H, Sheppard AE. 2017. Plasmid classification in an era of whole-genome sequencing: application in studies of antibiotic resistance epidemiology. Front Microbiol 8:182. doi:10.3389/fmicb.2017.00182 - DOI - PMC - PubMed
-
- Weber RE, Pietsch M, Frühauf A, Pfeifer Y, Martin M, Luft D, Gatermann S, Pfennigwerth N, Kaase M, Werner G, Fuchs S. 2019. IS26-mediated transfer of blaNDM–1 as the main route of resistance transmission during a polyclonal, multispecies outbreak in a German hospital. Front Microbiol 10:1–14. doi:10.3389/fmicb.2019.02817 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

