χ-Conotoxins are an Evolutionary Innovation of Mollusk-Hunting Cone Snails as a Counter-Adaptation to Prey Defense
- PMID: 39470581
- PMCID: PMC11568388
- DOI: 10.1093/molbev/msae226
χ-Conotoxins are an Evolutionary Innovation of Mollusk-Hunting Cone Snails as a Counter-Adaptation to Prey Defense
Abstract
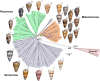
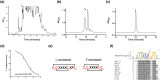
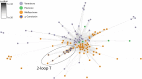
Mollusk-hunting (molluscivorous) cone snails belong to a monophyletic group in Conus, a genus of venomous marine snails. The molluscivorous lineage evolved from ancestral worm-hunting (vermivorous) snails ∼18 Ma. To enable the shift to a molluscivorous lifestyle, molluscivorous cone snails must solve biological problems encountered when hunting other gastropods, namely: (i) preventing prey escape and (ii) overcoming the formidable defense of the prey in the form of the molluscan shell, a problem unique to molluscivorous Conus. Here, we show that χ-conotoxins, peptides exclusively expressed in the venoms of molluscivorous Conus, provide solutions to the above problems. Injecting χ-conotoxins into the gastropod mollusk Aplysia californica results in impaired locomotion and uncoordinated hyperactivity. Impaired locomotion impedes escape, and a hyperactive snail will likely emerge from its shell, negating the protection the shell provides. Thus, χ-conotoxins are an evolutionary innovation that accompanied the emergence of molluscivory in Conus and provide solutions to problems posed by hunting other snails.
Keywords: conotoxins; emergence of complex phenotypes; evolutionary innovation; mollusk-hunting cone snails; χ-conotoxin.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest: The authors declare no conflict of interest in the conduct of this research.
Figures

References
-
- Abalde S, Dutertre S, Zardoya R. A combined transcriptomics and proteomics approach reveals the differences in the predatory and defensive venoms of the molluscivorous cone snail Cylinder ammiralis (Caenogastropoda: Conidae). Toxins (Basel). 2021:13(9):642. 10.3390/toxins13090642. - DOI - PMC - PubMed
-
- Abalde S, Tenorio MJ, Uribe JE, Zardoya R. Conidae phylogenomics and evolution. Zool Scr. 2019:48(2):194–214. 10.1111/zsc.12329. - DOI
-
- Adamo SA. Norepinephrine and octopamine: linking stress and immune function across phyla. Invert Surv J. 2008:5:12–19. 10.1093/icb/icu005. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

