IMPA1-derived inositol maintains stemness in castration-resistant prostate cancer via IMPDH2 activation
- PMID: 39470689
- PMCID: PMC11528126
- DOI: 10.1084/jem.20231832
IMPA1-derived inositol maintains stemness in castration-resistant prostate cancer via IMPDH2 activation
Abstract
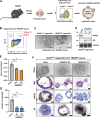
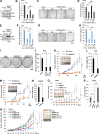
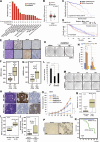
Acquisition of prostate cancer stem cells (PCSCs) manifested during androgen ablation therapy (ABT) contributes to castration-resistant prostate cancer (CRPC). However, little is known about the specific metabolites critically orchestrating this process. Here, we show that IMPA1-derived inositol enriched in PCSCs is a key metabolite crucially maintaining PCSCs for CRPC progression and ABT resistance. Notably, conditional Impa1 knockout in the prostate abrogates the pool and properties of PCSCs to orchestrate CRPC progression and prolong the survival of TRAMP mice. IMPA1-derived inositol serves as a cofactor that directly binds to and activates IMPDH2, which synthesizes guanylate nucleotides for maintaining PCSCs with ARlow/- features leading to CRPC progression and ABT resistance. IMPA1/inositol/IMPDH2 axis is upregulated in human prostate cancer, and its overexpression predicts poor survival outcomes. Genetically and pharmacologically targeting the IMPA1/inositol/IMPDH2 axis abrogates CRPC and overcomes ABT resistance in various CRPC xenografts, patient-derived xenograft (PDX) tumor models, and TRAMP mouse models. Our study identifies IMPDH2 as an inositol sensor whose activation by inositol represents a key mechanism for maintaining PCSCs for CRPC and ABT resistance.
© 2024 Hsu et al.
Conflict of interest statement
Disclosures: A.J. Armstrong reported personal fees from Pfizer/Astellas, personal fees from AstraZeneca, personal fees from Bayer, personal fees from Novartis, grants from BMS, and personal fees from Merck during the conduct of the study. J. Huang reported personal fees from Kingmed Diagnostics, personal fees from Artera, and personal fees from York Biotechnology outside the submitted work. H.-K. Lin reported personal fees from Stablix, Inc. outside the submitted work. No other disclosures were reported.
Figures













References
-
- Aggarwal, R., Huang J., Alumkal J.J., Zhang L., Feng F.Y., Thomas G.V., Weinstein A.S., Friedl V., Zhang C., Witte O.N., et al. . 2018. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: A multi-institutional prospective study. J. Clin. Oncol. 36:2492–2503. 10.1200/JCO.2017.77.6880 - DOI - PMC - PubMed
-
- Beltran, H., Rickman D.S., Park K., Chae S.S., Sboner A., MacDonald T.Y., Wang Y., Sheikh K.L., Terry S., Tagawa S.T., et al. . 2011. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 1:487–495. 10.1158/2159-8290.CD-11-0130 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

