BacDive in 2025: the core database for prokaryotic strain data
- PMID: 39470737
- PMCID: PMC11701647
- DOI: 10.1093/nar/gkae959
BacDive in 2025: the core database for prokaryotic strain data
Abstract
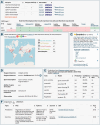
In 2025, the bacterial diversity database BacDive is the leading database for strain-level bacterial and archaeal information. It has been selected as an ELIXIR Core Data Resource as well as a Global Core Biodata Resource. Since its initial release more than ten years ago, BacDive (https://bacdive.dsmz.de) has grown tremendously in content and functionalities, and is a comprehensive resource covering the phenotypic diversity of prokaryotes with data on taxonomy, morphology, physiology, cultivation, and more. The current release (2023.2) contains 2.6 million data points on 97 334 strains, reflecting an increase by 52% since the previous publication in 2021. This remarkable growth can largely be attributed to the integration of the world-wide largest collection of Analytical Profile Index (API) test results, which are now fully integrated into the database and searchable. A novel BacDive knowledge graph provides powerful search options through a SPARQL endpoint, including the possibility for federated searches across multiple data sources. The high-quality data provided by BacDive is increasingly being used for the training of artificial intelligence models and resulting genome-based predictions with high confidence are now used to fill content gaps in the database.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Palma T.L., Costa M.C.. Biodegradation of 17α-ethinylestradiol by strains of Aeromonas genus isolated from acid mine drainage. Clean Technol. 2024; 6:116–139.
-
- Kapoor A., Varshney C.. Microbial degradation of PET plastic sustainably yielding commercially viable products. 2021; Preprints doi:21 June 2021, pre-print: not peer-reviewed10.20944/preprints202106.0519.v1. - DOI
-
- da Silva Santos D., Freitas N.S.A., de Morais M.A., Mendonça A.A.. Liquorilactobacillus: a context of the evolutionary history and metabolic adaptation of a bacterial genus from fermentation liquid environments. J. Mol. Evol. 2024; 92:467–487. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

