Comprehensive analysis of mesenchymal cells reveals a dysregulated TGF-β/WNT/HOXB7 axis in patients with myelofibrosis
- PMID: 39470742
- PMCID: PMC11623938
- DOI: 10.1172/jci.insight.173665
Comprehensive analysis of mesenchymal cells reveals a dysregulated TGF-β/WNT/HOXB7 axis in patients with myelofibrosis
Abstract
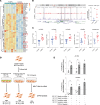
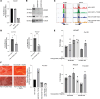
Despite the advances in the understanding and treatment of myeloproliferative neoplasm (MPN), the disease remains incurable with the risk of evolution to acute myeloid leukemia or myelofibrosis (MF). Unfortunately, the evolution of the disease to MF remains poorly understood, impeding preventive and therapeutic options. Recent studies in solid tumor microenvironment and organ fibrosis have shed instrumental insights on their respective pathogenesis and drug resistance, yet such precise data are lacking in MPN. In this study, through a patient sample-driven transcriptomic and epigenetic description of the MF microenvironment landscape and cell-based analyses, we identify homeobox B7 (HOXB7) overexpression and more precisely a potentially novel TGF-β/WNT/HOXB7 pathway as associated to a pro-fibrotic and pro-osteoblastic biased differentiation of mesenchymal stromal cells (MSCs). Using gene-based and chemical inhibition of this pathway, we reversed the abnormal phenotype of MSCs from patients with MF, providing the MPN field a potentially novel target to prevent and manage evolution to MF.
Keywords: Bone marrow; Fibrosis; Hematology.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

