Genetic associations of neuropathic pain and sensory profile in a deeply phenotyped neuropathy cohort
- PMID: 39471050
- PMCID: PMC12067614
- DOI: 10.1097/j.pain.0000000000003463
Genetic associations of neuropathic pain and sensory profile in a deeply phenotyped neuropathy cohort
Abstract
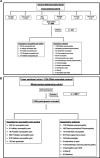
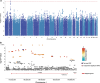
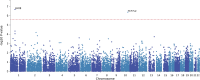
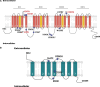
We aimed to investigate the genetic associations of neuropathic pain in a deeply phenotyped cohort. Participants with neuropathic pain were cases and compared with those exposed to injury or disease but without neuropathic pain as control subjects. Diabetic polyneuropathy was the most common aetiology of neuropathic pain. A standardised quantitative sensory testing protocol was used to categorize participants based on sensory profile. We performed genome-wide association study, and in a subset of participants, we undertook whole-exome sequencing targeting analyses of 45 known pain-related genes. In the genome-wide association study of diabetic neuropathy (N = 1541), a top significant association was found at the KCNT2 locus linked with pain intensity (rs114159097, P = 3.55 × 10 -8 ). Gene-based analysis revealed significant associations between LHX8 and TCF7L2 and neuropathic pain. Polygenic risk score for depression was associated with neuropathic pain in all participants. Polygenic risk score for C-reactive protein showed a positive association, while that for fasting insulin showed a negative association with neuropathic pain, in individuals with diabetic polyneuropathy. Gene burden analysis of candidate pain genes supported significant associations between rare variants in SCN9A and OPRM1 and neuropathic pain. Comparison of individuals with the "irritable" nociceptor profile to those with a "nonirritable" nociceptor profile identified a significantly associated variant (rs72669682, P = 4.39 × 10 -8 ) within the ANK2 gene. Our study on a deeply phenotyped cohort with neuropathic pain has confirmed genetic associations with the known pain-related genes KCNT2 , OPRM1 , and SCN9A and identified novel associations with LHX8 and ANK2 , genes not previously linked to pain and sensory profiles, respectively.
Keywords: Diabetes mellitus; GWAS; Neuropathic pain; Neuropathy; Sensory profile.
Copyright © 2024 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the International Association for the Study of Pain.
Conflict of interest statement
D. L. Bennett has acted as a consultant in the last 2 years for AditumBio, Amgen, Biogen, Biointervene, Combigene, LatigoBio, GSK, Ionis, Lexicon therapeutics, Lilly, Neuvati, Novo Ventures, Orion, Replay, SC Health Managers, Third Rock ventures, Vida Ventures on behalf of Oxford University Innovation. He has received research funding from Eli Lilly and Astra Zeneca. He has received an industrial partnership grant from the BBSRC and AstraZeneca. B. Smith has received research funding from Eli Lilly. N. Attal has received consultancy fees or participated as speaker bureau in the last 2 years for Merz, Grunenthal, Biogen, Novartis, Medtronic, Pfizer and Viatris outside the submitted work. N. B. Finnerup has acted as consultant for PharmNovo, Vertex, NeuroPN, Saniona, Nanobiotix, Neurvati, Biogen, Merz, and Confo Therapeutics. She has received grants from IMI2PainCare an EU IMI 2 (Innovative medicines initiative) public–private consortium, and the companies involved are: Grunenthal, Bayer, Eli Lilly, Esteve, and Teva, outside the submitted work. D. Bouhassira has received consultancy fees from Grunenthal and Bayer in the last 2 years. N. van Zuydam is currently an employee of AstraZeneca and a shareholder of AstraZeneca stock. R. Baron is supported by the EUROPAIN project, which is a public–private partnership and has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115007, resources for which are composed of financial contribution from the European Union's Seventh Framework Programme (FP7/2007-2013) and European Federation of Pharmaceutical Industries and Associations (EFPIA) companies' in-kind contribution. The NEUROPAIN project is an investigator-initiated European multicentre study with R. Baron as the principal investigator and 10 co-investigator sites, supported by an independent investigator-initiated research grant from Pfizer Ltd. R. Baron has also received Research grant funding from: Pfizer Pharma GmbH, Grünenthal GmbH, Mundipharma Research GmbH und Co. KG., Alnylam Pharmaceuticals Inc., Zambon GmbH, Sanofi Aventis GmbH, Viatris. The funding source had no role in study design, data collection and analysis, or writing of the manuscript. Outside the submitted word, J. Gierthmühlen has received consultancy fees from TEVA and Omega Pharma. She has received grants from companies: ElectroZeutica, Bosana GmBh, and Neurotech GmbH and personal fees for lectures from Teva, Abbvie, Lilly GmbH, Lundbeck, Grünenthal, CHangePain, StreamUp, MediSage, CampusWebinar. A. S.C. Rice interests occurring in last 24 months: Officer (President-Elect) of International Association for the Study of Pain; ASCR undertakes consultancy and advisory board work for Imperial College Consultants in the last 24 months this has included remunerated work for: AstraZeneca, Pharmnovo, Confo and Combigene. A. S.C. Rice is named as an inventor on patents: Rice ASC, Vandevoorde S, Lambert D.M Methods using N-(2-propenyl) hexadecanamide and related amides to relieve pain. WO 2005/079771, Okuse K. et al. Methods of treating pain by inhibition of vgf activity EP13702262.0/WO2013 110945. Member Joint Committee on Vaccine and Immunisation- varicella sub-committee; Analgesic Clinical Trial Translation: Innovations, Opportunities, and Networks (ACTTION) steering committee member; Medicines and Healthcare products Regulatory Agency (MHRA), Commission on Human Medicines—Neurology, Pain & Psychiatry Expert Advisory Group. Grants and studentships—UKRI (Medical Research Council & BBSRC), Versus Arthritis, Alan and Sheila Diamond Trust, Royal British Legion, European Commission, Ministry of Defence, Dr. Jennie Gwynn Bequests, The British Pain Society, Royal Society of Medicine. R. Baron has acted as a consultant for Pfizer Pharma GmbH, Sanofi Aventis GmbH, Grünenthal GmbH, Lilly, Novartis Pharma GmbH, Bristol-Myers Squibb, Biogenidec, AstraZeneca GmbH, Daiichi Sankyo, Glenmark Pharmaceuticals S.A., Seqirus Australia Pty Ltd, Teva Pharmaceuticals Europe Niederlande, Teva GmbH, Genentech, Mundipharma International Ltd UK, Galapagos NV, Kyowa Kirin GmbH, Vertex Pharmaceuticals Inc, Biotest AG, Celgene GmbH, Desitin Arzneimittel GmbH, Regeneron Pharmaceuticals Inc USA, Theranexus DSV CEA Frankreich, Abbott Products Operations AG Schweiz, Bayer AG, Grünenthal Pharma AG Schweiz, Akcea Therapeutics Germany GmbH, Asahi Kasei Pharma Corporation, AbbVie Deutschland GmbH & Co KG, Air Liquide Sante International Frankreich, Alnylam Germany GmbH, Lateral Pharma Pty Ltd, Hexal AG, Angelini, Janssen, SIMR Biotech Pty Ltd Australien, Confo Therapeutics N. V. Belgium, Merz Pharmaceuticals GmbH, Neumentum Inc, F. Hoffmann-La Roche Ltd Switzerland, AlgoTherapeutix SAS France, Nanobiotix SA France, AmacaThera Inc Canada, Heat2Move, Resano GmbH, Esteve Pharmaceuticals SA. R. Baron has acted as a Speaker for Pfizer Pharma GmbH, Sanofi Aventis GmbH, Grünenthal GmbH, Mundipharma, Lilly GmbH, Desitin Arzneimittel GmbH, Teva GmbH, Bayer AG, MSD GmbH, Seqirus Australia Pty Ltd, Novartis Pharma GmbH, TAD Pharma GmbH, Grünenthal SA Portugal, Grünenthal Pharma AG Schweiz, Grünenthal B.V. Niederlande, Evapharma, Takeda Pharmaceuticals International AG Schweiz, Ology Medical Education Netherlands, Ever Pharma GmbH, Amicus Therapeutics GmbH, Novo Nordisk Pharma GmbH, Chiesi GmbH, Stada Mena DWC LLC Dubai, Hexal AG, Viatris, AstraZeneca GmbH, Sandoz.
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Figures
References
-
- Almomani R, Sopacua M, Marchi M, Ślęczkowska M, Lindsey P, de Greef BT, Hoeijmakers JG, Salvi E, Merkies IS, Ferdousi M, Malik RA, Ziegler D, Derks KWJ, Boenhof G, Martinelli-Boneschi F, Cazzato D, Lombardi R, Dib-Hajj S, Waxman SG, Smeets HJM, Gerrits MM, Faber CG, Lauria G; On Behalf Of The Propane Study Group. Genetic profiling of sodium channels in diabetic painful and painless and idiopathic painful and painless neuropathies. Int J Mol Sci 2023;24:8278. - PMC - PubMed
-
- Attal N, Lanteri-Minet M, Laurent B, Fermanian J, Bouhassira D. The specific disease burden of neuropathic pain: results of a French nationwide survey. PAIN 2011;152:2836–43. - PubMed
-
- Baron R, Dickenson AH, Calvo M, Dib-Hajj SD, Bennett DL. Maximizing treatment efficacy through patient stratification in neuropathic pain trials. Nat Rev Neurol 2023;19:53–64. - PubMed
-
- Baron R, Maier C, Attal N, Binder A, Bouhassira D, Cruccu G, Finnerup NB, Haanpää M, Hansson P, Hüllemann P, Jensen TS, Freynhagen R, Kennedy JD, Magerl W, Mainka T, Reimer M, Rice ASC, Segerdahl M, Serra J, Sindrup S, Sommer C, Tölle T, Vollert J, Treede RD; German Neuropathic Pain Research Network DFNS, and the EUROPAIN, and NEUROPAIN consortia. Peripheral neuropathic pain: a mechanism-related organizing principle based on sensory profiles. PAIN 2017;158:261–72. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

