Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity
- PMID: 39471218
- PMCID: PMC11551396
- DOI: 10.1073/pnas.2417144121
Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity
Abstract
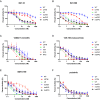
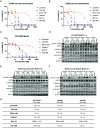
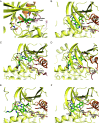
Somatic mutations in the epidermal growth factor receptor (EGFR) are a major cause of non-small cell lung cancer. Among these structurally diverse alterations, exon 20 insertions represent a unique subset that rarely respond to EGFR tyrosine kinase inhibitors (TKIs). Therefore, there is a significant need to develop inhibitors that are active against this class of activating mutations. Here, we conducted biochemical analysis of the two most frequent exon 20 insertion variants, V769_D770insASV (insASV) and D770_N771insSVD (insSVD) to better understand their drug sensitivity and resistance. From kinetic studies, we found that EGFR insASV and insSVD are similarly active, but have lower Km, ATP values compared to the L858R variant, which contributes to their lack of sensitivity to 1st-3rd generation EGFR TKIs. Biochemical, structural, and cellular studies of a diverse panel of EGFR inhibitors revealed that the more recently developed compounds BAY-568, TAS6417, and TAK-788 inhibit EGFR insASV and insSVD in a mutant-selective manner, with BAY-568 being the most potent and selective versus wild-type (WT) EGFR. Cocrystal structures with WT EGFR reveal the binding modes of each of these inhibitors and of poziotinib, a potent but not mutantselective inhibitor, and together they define interactions shared by the mutant-selective agents. Collectively, our results show that these exon20 insertion variants are not inherently inhibitor resistant, rather they differ in their drug sensitivity from WT EGFR. However, they are similar to each other, indicating that a single inhibitor should be effective for several of the diverse exon 20 insertion variants.
Keywords: EGFR inhibitors; X-ray crystallography; enzymology; lung cancer.
Conflict of interest statement
Competing interests statement:M.J.E. receives or has received sponsored research support from Novartis, Sanofi, Takeda, and Springworks Therapeutics and consulting income or honoraria from Novartis, H3 Biomedicine and Ikena Oncology. P.A.J. has received consulting fees from AstraZeneca, Boehringer Ingelheim, Pfizer, Roche/Genentech, Takeda Oncology, ACEA Biosciences, Eli Lilly and Company, Araxes Pharma, Ignyta, Mirati Therapeutics, Novartis, LOXO Oncology, Daiichi Sankyo, Sanofi Oncology, Voronoi, SFJ Pharmaceuticals, Biocartis, Novartis Oncology, Nuvalent, Esai, Bayer, Transcenta, Silicon Therapeutics, Allorion Therapeutics, Accutar Biotech and AbbVie; receives post-marketing royalties from DFCI-owned intellectual property on EGFR mutations licensed to Lab Corp; receives or has received sponsored research funding from AstraZeneca, Astellas, Daichi-Sankyo, PUMA, Boehringer Ingelheim, Eli Lilly and Company, Revolution Medicines and Takeda and has stock ownership in Gatekeeper Pharmaceuticals. H.Z. was a consultant at Boston Consulting Group and is currently a consultant at Bain & Company. All other authors declare no competing interests.
Figures

References
-
- Sung H., et al. , Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). - PubMed
-
- Siegel R. L., Miller K. D., Fuchs H. E., Jemal A., Cancer statistics. CA Cancer J. Clin. 71, 7–33 (2021). - PubMed
-
- Lynch T. J., et al. , Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139 (2004). - PubMed
-
- Paez J. G., et al. , EGFR mutations in lung, cancer: Correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004). - PubMed
-
- Sordella R., Bell D. W., Haber D. A., Settleman J., Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science 305, 1163–1167 (2004). - PubMed
MeSH terms
Substances
Grants and funding
- CA242461/HHS | NIH | National Cancer Institute (NCI)
- P41 GM103403/GM/NIGMS NIH HHS/United States
- CA154303/HHS | NIH | National Cancer Institute (NCI)
- F32CA247198/HHS | NIH | National Cancer Institute (NCI)
- CA220497/HHS | NIH | National Cancer Institute (NCI)
- R35 CA242461/CA/NCI NIH HHS/United States
- CA201049/HHS | NIH | National Cancer Institute (NCI)
- P01 CA154303/CA/NCI NIH HHS/United States
- S10 OD021527/OD/NIH HHS/United States
- F32 CA247198/CA/NCI NIH HHS/United States
- R01 CA201049/CA/NCI NIH HHS/United States
- P30 GM124165/GM/NIGMS NIH HHS/United States
- R35 CA220497/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

