Epigenetic features support the diagnosis of B-cell prolymphocytic leukemia and identify 2 clinicobiological subtypes
- PMID: 39471431
- PMCID: PMC11700275
- DOI: 10.1182/bloodadvances.2024013327
Epigenetic features support the diagnosis of B-cell prolymphocytic leukemia and identify 2 clinicobiological subtypes
Abstract
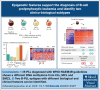
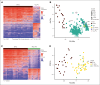
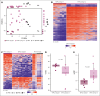
The recognition of B-cell prolymphocytic leukemia (B-PLL) as a separate entity is controversial based on the current classification systems. Here, we analyzed the DNA methylome of a cohort of 20 B-PLL cases diagnosed according to the guidelines of the International Consensus Classification/Fourth revised edition of the World Health Organization Classification, and compared them with chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL), splenic marginal zone lymphoma (SMZL), and normal B-cell subpopulations. Unsupervised principal component analyses suggest that B-PLL is epigenetically distinct from CLL, MCL, and SMZL, which is further supported by robust differential methylation signatures in B-PLL. We also observe that B-PLL can be segregated into 2 epitypes with differential clinicobiological characteristics. B-PLL epitype 1 carries lower immunoglobulin heavy variable somatic hypermutation and a less profound germinal center-related DNA methylation imprint than epitype 2. Furthermore, epitype 1 is significantly enriched in mutations affecting MYC and SF3B1, and displays DNA hypomethylation and gene upregulation signatures enriched in MYC targets. Despite the low sample size, patients from epitype 1 have an inferior overall survival than those of epitype 2. This study provides relevant insights into the biology and differential diagnosis of B-PLL, and potentially identifies 2 subgroups with distinct biological and clinical features.
© 2024 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures


References
-
- Catovsky D, Galetto J, Okos A, et al. Prolymphocytic leukaemia of B and T cell type. Lancet. 1973;(2):233–234. - PubMed
-
- Galton DAG, Goldman JM, Wiltshaw E, Catovsky D, Henry K, Goldenberg GJ. Prolymphocytic leukaemia. Br J Haematol. 1974;27(1):7–23. - PubMed
-
- Catovsky D, Montserrat E, Muller-Hermelink HK, Harris NL. In: WHO Classification of Tumours - Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissue. Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. IARC Press; 2001. B-cell prolymphocytic leukemia; pp. 131–132.
-
- Chapiro E, Pramil E, Diop M, et al. Genetic characterization of B-cell prolymphocytic leukemia: a prognostic model involving MYC and TP53. Blood. 2019;134(21):1821–1831. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

