LGR6 protects against myocardial ischemia-reperfusion injury via suppressing necroptosis
- PMID: 39471639
- PMCID: PMC11550357
- DOI: 10.1016/j.redox.2024.103400
LGR6 protects against myocardial ischemia-reperfusion injury via suppressing necroptosis
Abstract
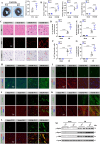
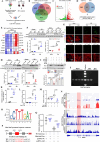
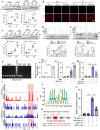
Regulated necrosis (necroptosis) and apoptosis are important biological features of ischemia-reperfusion (I/R) injury. However, the molecular mechanisms underlying myocardial necroptosis remain elusive. Leucine rich repeat containing G protein-coupled receptor 6 (LGR6) has been reported to play important roles in various cardiovascular disease. In this study, we aimed to determine whether LGR6 suppresses I/R-induced myocardial necroptosis and the underlying molecular mechanisms. We generated LGR6 knockout mice and used ligation of left anterior descending coronary artery to produce an in vivo I/R model. The effects of LGR6 and its downstream molecules were subsequently identified using RNA sequencing and CHIP assays. We observed significantly downregulated LGR6 expression in hearts post myocardial I/R and cardiomyocytes post hypoxia and reoxygenation (HR). LGR6 deficiency promoted and LGR6 overexpression inhibited necroptosis and acute myocardial injury after I/R. Mechanistically, in vivo and in vitro experiments suggest that LGR6 regulates the expression of STAT2 and ZBP1 by activating the Wnt signaling pathway, thereby inhibiting cardiomyocyte necroptosis after HR. Inhibiting STAT2 and ZBP1 effectively alleviated the aggravating effect of LGR6 deficiency on myocardial necroptosis after I/R. Furthermore, activating LGR6 with RSPO3 also effectively protected mice from acute myocardial I/R injury. Our findings reveal that RSPO3-LGR6 axis downregulates the expression of STAT2 and ZBP1 through the Wnt signaling pathway, thereby inhibiting I/R-induced myocardial injury and necroptosis. Targeting the RSPO3-LGR6 axis may be a potential therapeutic strategy to treat myocardial I/R injury.
Keywords: LGR6; Myocardial ischemia-reperfusion injury; Necroptosis; STAT2; ZBP1.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

