Predictive gene expression signature diagnoses neonatal sepsis before clinical presentation
- PMID: 39472236
- PMCID: PMC11663757
- DOI: 10.1016/j.ebiom.2024.105411
Predictive gene expression signature diagnoses neonatal sepsis before clinical presentation
Abstract
Background: Neonatal sepsis is a deadly disease with non-specific clinical signs, delaying diagnosis and treatment. There remains a need for early biomarkers to facilitate timely intervention. Our objective was to identify neonatal sepsis gene expression biomarkers that could predict sepsis at birth, prior to clinical presentation.
Methods: Among 720 initially healthy full-term neonates in two hospitals (The Gambia, West Africa), we identified 21 newborns who were later hospitalized for sepsis in the first 28 days of life, split into early-onset sepsis (EOS, onset ≤7 days of life) and late-onset sepsis (LOS, onset 8-28 days of life), 12 neonates later hospitalized for localized infection without evidence of systemic involvement, and 33 matched control neonates who remained healthy. RNA-seq was performed on peripheral blood collected at birth when all neonates were healthy and also within the first week of life to identify differentially expressed genes (DEGs). Machine learning methods (sPLS-DA, LASSO) identified genes expressed at birth that predicted onset of neonatal sepsis at a later time.
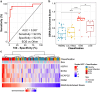
Findings: Neonates who later developed EOS already had ∼1000 DEGs at birth when compared to control neonates or those who later developed a localized infection or LOS. Based on these DEGs, a 4-gene signature (HSPH1, BORA, NCAPG2, PRIM1) for predicting EOS at birth was developed (training AUC = 0.94, sensitivity = 0.93, specificity = 0.92) and validated in an external cohort (validation AUC = 0.72, sensitivity = 0.83, and specificity = 0.83). Additionally, during the first week of life, EOS disrupted expression of >1800 genes including those influencing immune and metabolic transitions observed in healthy controls.
Interpretation: Despite appearing healthy at birth, neonates who later developed EOS already had distinct whole blood gene expression changes at birth, which enabled the development of a 4-gene predictive signature for EOS. This could facilitate early recognition and treatment of neonatal sepsis, potentially mitigating its long-term sequelae.
Funding: CIHR and NIH/NIAID.
Keywords: Machine learning; Neonatal sepsis; Ontogeny; Predictive biomarkers; Transcriptomics.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests AA provided legal consultation for MCIC Vermont Inc and received honorarium for lecture on neonatal sepsis from the University of Rome. BK participated in a Data Safety Monitoring Board for Johnson & Johnson. JDA received travel funding from the American Society for Histocompatibility and Immunogenetics, World Vaccine Congress, and International Network of Special Immunization Services. OAO received travel funding from the American Academy of Pediatrics. OL is a named inventor on patents held by Boston Children's Hospital relating to small molecule adjuvants (e.g., Novel imidazopyrimidine compounds and uses thereof; EP3709998A1) and to human in vitro systems that model responses to immunomodulators and vaccines (e.g., Tissue constructs and uses thereof; US20150152385A1), has served as a consultant to Glaxo Smith Kline and Hillevax, and is co-founder of and advisor to Ovax, Inc. REWH has a contract from Sepset Biosciences for development of diagnostic assays for adult sepsis (indirect relationship to this work) and is CEO of Asep Medical and Sepset BioSciences that are commercially developing adult sepsis diagnostics, although the signatures described here have not been filed for patent protection. All other authors declare no conflict of interest.
Figures



References
-
- Okomo U., Akpalu E.N.K., Le Doare K., et al. Aetiology of invasive bacterial infection and antimicrobial resistance in neonates in sub-Saharan Africa: a systematic review and meta-analysis in line with the STROBE-NI reporting guidelines. Lancet Infect Dis. 2019;19(11):1219–1234. doi: 10.1016/S1473-3099(19)30414-1. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

