Gut microbial and human genetic signatures of inflammatory bowel disease increase risk of comorbid mental disorders
- PMID: 39472439
- PMCID: PMC11522461
- DOI: 10.1038/s41525-024-00440-w
Gut microbial and human genetic signatures of inflammatory bowel disease increase risk of comorbid mental disorders
Abstract
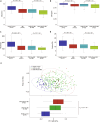
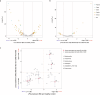
The high prevalence of comorbid mental disorders (CMDs) in patients with inflammatory bowel disease (IBD) is well-documented. This study delves into the intricate CMD-IBD relationship through comprehensive analyses using human variants, gut microbiome, and anxiety/depression estimates from a cohort of 507 IBD patients and 75 controls. Notably, patients with IBD, especially those with CMD, exhibited lower diversity than controls. We identified 106 differentially abundant taxa (DATs) in IBD patients compared to controls and 21 DATs distinguishing CMD-affected from CMD-free IBD patients. Microbial IBD-risk scores, reflecting an individual's microbial burden for IBD, revealed a significant enrichment of IBD-risk signatures in CMD-affected patients compared to CMD-free patients. Additionally, there was an IBD-risk variant potentially regulating the abundance of an IBD/CMD-associated DAT, suggesting an interplay between IBD-risk variants and dysbiosis in CMD. Our investigation underscores the pivotal role of IBD-associated gut dysbiosis in predisposing IBD patients to CMD, partially through genetic variant-mediated mechanisms.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

