Discovery and prioritization of genetic determinants of kidney function in 297,355 individuals from Taiwan and Japan
- PMID: 39472450
- PMCID: PMC11522641
- DOI: 10.1038/s41467-024-53516-7
Discovery and prioritization of genetic determinants of kidney function in 297,355 individuals from Taiwan and Japan
Abstract
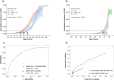
Current genome-wide association studies (GWAS) for kidney function lack ancestral diversity, limiting the applicability to broader populations. The East-Asian population is especially under-represented, despite having the highest global burden of end-stage kidney disease. We conducted a meta-analysis of multiple GWASs (n = 244,952) on estimated glomerular filtration rate and a replication dataset (n = 27,058) from Taiwan and Japan. This study identified 111 lead SNPs in 97 genomic risk loci. Functional enrichment analyses revealed that variants associated with F12 gene and a missense mutation in ABCG2 may contribute to chronic kidney disease (CKD) through influencing inflammation, coagulation, and urate metabolism pathways. In independent cohorts from Taiwan (n = 25,345) and the United Kingdom (n = 260,245), polygenic risk scores (PRSs) for CKD significantly stratified the risk of CKD (p < 0.0001). Further research is required to evaluate the clinical effectiveness of PRSCKD in the early prevention of kidney disease.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Savage, N. Tapping into the drug discovery potential of AI. Biopharm. Deal. B37–B39 https://www.nature.com/articles/d43747-021-00045-7 (2021).
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

