Structural basis for the transmembrane signaling and antidepressant-induced activation of the receptor tyrosine kinase TrkB
- PMID: 39472452
- PMCID: PMC11522581
- DOI: 10.1038/s41467-024-53710-7
Structural basis for the transmembrane signaling and antidepressant-induced activation of the receptor tyrosine kinase TrkB
Abstract
Neurotrophin receptors of the Trk family are involved in the regulation of brain development and neuroplasticity, and therefore can serve as targets for anti-cancer and stroke-recovery drugs, antidepressants, and many others. The structures of Trk protein domains in various states upon activation need to be elucidated to allow rational drug design. However, little is known about the conformations of the transmembrane and juxtamembrane domains of Trk receptors. In the present study, we employ NMR spectroscopy to solve the structure of the TrkB dimeric transmembrane domain in the lipid environment. We verify the structure using mutagenesis and confirm that the conformation corresponds to the active state of the receptor. Subsequent study of TrkB interaction with the antidepressant drug fluoxetine, and the antipsychotic drug chlorpromazine, provides a clear self-consistent model, describing the mechanism by which fluoxetine activates the receptor by binding to its transmembrane domain.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests
Figures





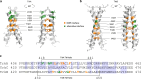
References
-
- Friedman, W. J. & Greene, L. A. Neurotrophin signaling via Trks and p75. Exp. Cell Res.253, 131–142 (1999). - PubMed
-
- Martin-Zanca, D., Hughes, S. H. & Barbacid, M. A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature319, 743–748 (1986). - PubMed
-
- Segal, R. A. Selectivity in neurotrophin signaling: theme and variations. Annu. Rev. Neurosci.26, 299–330 (2003). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- 22-14-00130/Russian Science Foundation (RSF)
- INVEST/2022/456/Regional Government of Valencia | Conselleria d'Educació, Investigació, Cultura i Esport (Conselleria d'Educació, Investigació, Cultura i Esport de la Generalitat Valenciana)
- CRC 1507/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Medical

