Two ubiquitous aldo-keto reductases in the genus Papaver support a patchwork model for morphine pathway evolution
- PMID: 39472466
- PMCID: PMC11522673
- DOI: 10.1038/s42003-024-07100-w
Two ubiquitous aldo-keto reductases in the genus Papaver support a patchwork model for morphine pathway evolution
Abstract
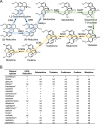
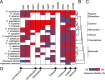
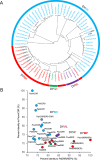
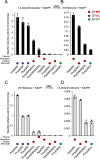
The evolution of morphinan alkaloid biosynthesis in plants of the genus Papaver includes permutation of several processes including gene duplication, fusion, neofunctionalization, and deletion resulting in the present chemotaxonomy. A critical gene fusion event resulting in the key bifunctional enzyme reticuline epimerase (REPI), which catalyzes the stereochemical inversion of (S)-reticuline, was suggested to precede neofunctionalization of downstream enzymes leading to morphine biosynthesis in opium poppy (Papaver somniferum). The ancestrally related aldo-keto reductases 1,2-dehydroreticuline reductase (DRR), which occurs in some species as a component of REPI, and codeinone reductase (COR) catalyze the second and penultimate steps, respectively, in the pathway converting (S)-reticuline to morphine. Orthologs for each enzyme isolated from the transcriptomes of 12 Papaver species were shown to catalyze their respective reactions in species that capture states of the metabolic pathway prior to key evolutionary events, including the gene fusion event leading to REPI, thus suggesting a patchwork model for pathway evolution. Analysis of the structure and substrate preferences of DRR orthologs in comparison with COR orthologs revealed structure-function relationships underpinning the functional latency of DRR and COR orthologs in the genus Papaver, thus providing insights into the molecular events leading to the evolution of the pathway.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Dastmalchi, M., Park, M. R., Morris, J. S. & Facchini, P. Family portraits: the enzymes behind benzylisoquinoline alkaloid diversity. Phytochem. Rev.17, 249–277 (2018). - DOI
-
- World Health Organization. World health organization model list of essential medicines. Ment. Holist. Health.: Some Int. Perspect.21, 119–134 (2019).
-
- Board, I. N. C. Narcotic Drugs - Technical Report. (2019).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

