Microbial dynamics and pulmonary immune responses in COVID-19 secondary bacterial pneumonia
- PMID: 39472555
- PMCID: PMC11522429
- DOI: 10.1038/s41467-024-53566-x
Microbial dynamics and pulmonary immune responses in COVID-19 secondary bacterial pneumonia
Abstract
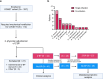
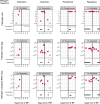
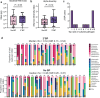
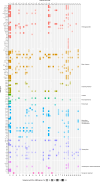
Secondary bacterial pneumonia (2°BP) is associated with significant morbidity following respiratory viral infection, yet remains incompletely understood. In a prospective cohort of 112 critically ill adults intubated for COVID-19, we comparatively assess longitudinal airway microbiome dynamics and the pulmonary transcriptome of patients who developed 2°BP versus controls who did not. We find that 2°BP is significantly associated with both mortality and corticosteroid treatment. The pulmonary microbiome in 2°BP is characterized by increased bacterial RNA mass and dominance of culture-confirmed pathogens, detectable days prior to 2°BP clinical diagnosis, and frequently also present in nasal swabs. Assessment of the pulmonary transcriptome reveals suppressed TNFα signaling in patients with 2°BP, and sensitivity analyses suggest this finding is mediated by corticosteroid treatment. Further, we find that increased bacterial RNA mass correlates with reduced expression of innate and adaptive immunity genes in both 2°BP patients and controls. Taken together, our findings provide fresh insights into the microbial dynamics and host immune features of COVID-19-associated 2°BP, and suggest that suppressed immune signaling, potentially mediated by corticosteroid treatment, permits expansion of opportunistic bacterial pathogens.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- BioProject/PRJNA1033689
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

