AAA+ ATPase chaperone p97/VCPFAF2 governs basal pexophagy
- PMID: 39472561
- PMCID: PMC11522385
- DOI: 10.1038/s41467-024-53558-x
AAA+ ATPase chaperone p97/VCPFAF2 governs basal pexophagy
Abstract
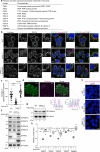
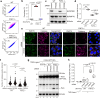
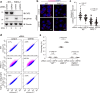
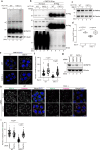
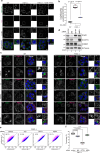
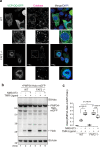
Peroxisomes are organelles that are central to lipid metabolism and chemical detoxification. Despite advances in our understanding of peroxisome biogenesis, the mechanisms maintaining peroxisomal membrane proteins remain to be fully elucidated. We show here that mammalian FAF2/UBXD8, a membrane-associated cofactor of p97/VCP, maintains peroxisomal homeostasis by modulating the turnover of peroxisomal membrane proteins such as PMP70. In FAF2-deficient cells, PMP70 accumulation recruits the autophagy adaptor OPTN (Optineurin) to peroxisomes and promotes their autophagic clearance (pexophagy). Pexophagy is also induced by p97/VCP inhibition. FAF2 functions together with p97/VCP to negatively regulate pexophagy rather than as a factor for peroxisome biogenesis. Our results strongly suggest that p97/VCPFAF2-mediated extraction of ubiquitylated peroxisomal membrane proteins (e.g., PMP70) prevents peroxisomes from inducing nonessential autophagy under steady state conditions. These findings provide insight into molecular mechanisms underlying the regulation of peroxisomal integrity by p97/VCP and its associated cofactors.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- JP19H05712/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP15K19037, JP18K14708, JP21K06161/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP18H05500, JP16K18545, JP18K06237/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP26116007, JP15K21743, JP17H03675/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP26000014, JP19H00997/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources
Miscellaneous

