METTL3 confers oxaliplatin resistance through the activation of G6PD-enhanced pentose phosphate pathway in hepatocellular carcinoma
- PMID: 39472692
- PMCID: PMC11894169
- DOI: 10.1038/s41418-024-01406-2
METTL3 confers oxaliplatin resistance through the activation of G6PD-enhanced pentose phosphate pathway in hepatocellular carcinoma
Abstract
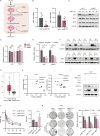
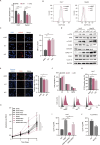
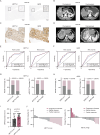
Oxaliplatin-based therapeutics is a widely used treatment approach for hepatocellular carcinoma (HCC) patients; however, drug resistance poses a significant clinical challenge. Epigenetic modifications have been implicated in the development of drug resistance. In our study, employing siRNA library screening, we identified that silencing the m6A writer METTL3 significantly enhanced the sensitivity to oxaliplatin in both in vivo and in vitro HCC models. Further investigations through combined RNA-seq and non-targeted metabolomics analysis revealed that silencing METTL3 impeded the pentose phosphate pathway (PPP), leading to a reduction in NADPH and nucleotide precursors. This disruption induced DNA damage, decreased DNA synthesis, and ultimately resulted in cell cycle arrest. Mechanistically, METTL3 was found to modify E3 ligase TRIM21 near the 3'UTR with N6-methyladenosine, leading to reduced RNA stability upon recognition by YTHDF2. TRIM21, in turn, facilitated the degradation of the rate-limiting enzyme of PPP, G6PD, through the ubiquitination-proteasome pathway. Importantly, high expression of METTL3 was significantly associated with adverse prognosis and oxaliplatin resistance in HCC patients. Notably, treatment with the specific METTL3 inhibitor, STM2457, significantly improved the efficacy of oxaliplatin. These findings underscore the critical role of the METTL3/TRIM21/G6PD axis in driving oxaliplatin resistance and present a promising strategy to overcome chemoresistance in HCC.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: All methods were performed in accordance with the relevant guidelines and regulations. All animal experiments were approved by the Animal Ethics Committee of Sun Yat-Sen University and performed in accordance with ethical requirements with the approval number L102012019005F. The usage of HCC tissue specimens was approved by the ethics committee of the Sun Yat-sen University Cancer Center, with informed consent obtained from all participants (Approval Number GZR2019-240).
Figures




References
-
- Sung H, Ferlay J, Siegel R, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin2021;71:209–49. - PubMed
-
- Lyu N, Wang X, Li J, Lai J, Chen Q, Li S, et al. Arterial chemotherapy of oxaliplatin plus fluorouracil versus sorafenib in advanced hepatocellular carcinoma: a biomolecular exploratory, randomized, phase III trial (FOHAIC-1). J Clin Oncol. 2022;40:468–80. - PubMed
-
- Lyu N, Kong Y, Mu L, Lin Y, Li J, Liu Y, et al. Hepatic arterial infusion of oxaliplatin plus fluorouracil/leucovorin vs. sorafenib for advanced hepatocellular carcinoma. J Hepatol. 2018;69:60–9. - PubMed
MeSH terms
Substances
Grants and funding
- 82172646/National Natural Science Foundation of China (National Science Foundation of China)
- 82202905/National Natural Science Foundation of China (National Science Foundation of China)
- 82173016/National Natural Science Foundation of China (National Science Foundation of China)
- 81972227/National Natural Science Foundation of China (National Science Foundation of China)
- 2021A1515010707/Natural Science Foundation of Guangdong Province (Guangdong Natural Science Foundation)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

