Olanzapine enhances early brain maturation through activation of the NODAL/FOXH1 axis
- PMID: 39474064
- PMCID: PMC11519431
- DOI: 10.1016/j.isci.2024.110917
Olanzapine enhances early brain maturation through activation of the NODAL/FOXH1 axis
Abstract
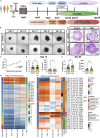
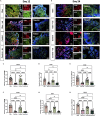
The portrayed effects of olanzapine on brain development and neuronal response remain unclear under the genetic background of Homo sapiens. Here, we constructed therapeutic-dosage olanzapine-treated cerebral organoid (CO) models using induced pluripotent stem cells from human samples. We found that the activation of NODAL/FOXH1 axis mediated the early response to olanzapine up to day 15, which subsequently caused thicker cortical-like structures, cell identity maturation, higher stemness of neural progenitor cells (NPCs), and mature neuronal firing of early neurons in day 24. Transcriptomics and targeted metabolomics confirmed the upregulation of neurodevelopmental-related terms and glutamate production on day 24. Gene enrichment of transcriptomics into large-scale genome-wide association studies (GWAS) showed possible relationships with intelligence, major depressive disorder, schizophrenia. We did not observe the negative effects of in-utero exposure to olanzapine in mice. Collectively, we tended to conclude that olanzapine treatment had beneficial effects instead of harmful on early brain development.
Keywords: Biological sciences; Cellular neuroscience; Developmental neuroscience; Natural sciences; Neuroscience.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Thomas K., Saadabadi A. StatPearls. 2023. Olanzapine.
LinkOut - more resources
Full Text Sources

