Regulation of BCR-dependent germinal center B-cell formation by HGAL and insight into its emerging myeloid ortholog, C1ORF150
- PMID: 39474423
- PMCID: PMC11518702
- DOI: 10.3389/fimmu.2024.1437516
Regulation of BCR-dependent germinal center B-cell formation by HGAL and insight into its emerging myeloid ortholog, C1ORF150
Abstract
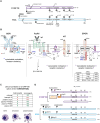
The specificity of cytokine and immunoreceptor signaling frequently depends upon receptor recruitment of select adaptor proteins and specifically engaged effectors. This review focuses on the orthologous adaptor proteins, HGAL and C1ORF150, and aims to provide insight into their respective modulation of lymphoid and myeloid cell signaling, formation, and function. HGAL acts predominantly within germinal center B cells as an important BCR signal transducer. Effects on BCR signalosome assembly involve HGAL's localization to the plasma membrane via its lipidation, initial interactions with SYK, the pY-phosphorylation of HGAL including its recruitment of GRB2, and HGAL engagement of PDZ-RhoGEF and RhoA signaling. At ligated BCRs, this includes HGAL(-GRB2) stimulation of SYK kinase, attenuation of calcium flux-dependent and NF-κB expression, promotion of cSMAC formation, and cytoskeletal remodeling associated with HGAL-attenuated cell migration. HGAL and partnered effectors also impact on DLBCL pathogenesis, and studies are summarized on HGAL's actions (using DLBCL and Burkitt lymphoma B cells) including cell migration effects, HGAL modulation of cytoskeletal components, and insightful HGAL transgenic mouse and xenograft models. For C1ORF150, its HGAL-homologous subdomains are considered, together with studies that demonstrate C1OR150's FcϵRI- and KIT-mediated expression and phosphorylation in primary human mast cells. Intriguingly, recent GWAS studies have identified a C1ORF150 in-frame splice variant that is strongly associated with urticaria. Candidate mechanisms via which the encoded "C1ORF150-Δexon2" isoform affects mast cell degranulation are considered, including FcϵR1 and/or KIT receptor connections, and candidate "myristoylation switch" mechanisms.
Keywords: B-cells; C1ORF150; HGAL; adaptor proteins; mast cells.
Copyright © 2024 Toran, Novelli, Lazor, Vachon and Wojchowski.
Conflict of interest statement
Author JL was employed by the company Boehringer Ingelheim Pharmaceuticals, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

