Cryo-EM structures of the zinc transporters ZnT3 and ZnT4 provide insights into their transport mechanisms
- PMID: 39474773
- PMCID: PMC11726163
- DOI: 10.1002/1873-3468.15047
Cryo-EM structures of the zinc transporters ZnT3 and ZnT4 provide insights into their transport mechanisms
Abstract
Zinc transporters (ZnTs) act as H+/Zn2+ antiporters, crucial for zinc homeostasis. Brain-specific ZnT3 expressed in synaptic vesicles transports Zn2+ from the cytosol into vesicles and is essential for neurotransmission, with ZnT3 dysfunction associated with neurological disorders. Ubiquitously expressed ZnT4 localized to lysosomes facilitates the Zn2+ efflux from the cytosol to lysosomes, mitigating the cell injury risk. Despite their importance, the structures and Zn2+ transport mechanisms remain unclear. We characterized the three-dimensional structures of human ZnT3 (inward-facing) and ZnT4 (outward-facing) using cryo-electron microscopy. By combining these structures, we assessed the conformational changes that could occur within the transmembrane domain during Zn2+ transport. Our results provide a structural basis for a more comprehensive understanding of the H+/Zn2+ exchange mechanisms exhibited by ZnTs.
Keywords: ZnT; cryo‐electron microscopy; membrane protein; three‐dimensional structure; zinc transporter.
© 2024 The Author(s). FEBS Letters published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Figures

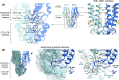


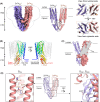

References
-
- Vallee BL and Falchuk KH (1993) The biochemical basis of zinc physiology. Physiol Rev 73, 79–118. - PubMed
-
- Maret W and Li Y (2009) Coordination dynamics of zinc in proteins. Chem Rev 109, 4682–4707. - PubMed
-
- Fukada T and Kambe T (2011) Molecular and genetic features of zinc transporters in physiology and pathogenesis. Metallomics 3, 662–674. - PubMed
MeSH terms
Substances
Grants and funding
- 20K16274/Grant-in-Aid from the Japanese Ministry of Education, Culture, Sports, Science, and Technology
- 22K07131/Grant-in-Aid from the Japanese Ministry of Education, Culture, Sports, Science, and Technology
- 22K15046/Grant-in-Aid from the Japanese Ministry of Education, Culture, Sports, Science, and Technology
- 24K0934/Grant-in-Aid from the Japanese Ministry of Education, Culture, Sports, Science, and Technology
LinkOut - more resources
Full Text Sources
Research Materials

