Single-Cell RNA Sequencing Uncovers Pathological Processes and Crucial Targets for Vascular Endothelial Injury in Diabetic Hearts
- PMID: 39475009
- PMCID: PMC11653609
- DOI: 10.1002/advs.202405543
Single-Cell RNA Sequencing Uncovers Pathological Processes and Crucial Targets for Vascular Endothelial Injury in Diabetic Hearts
Abstract
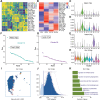
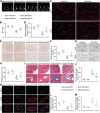
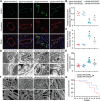
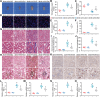
Cardiovascular disease remains the leading cause of high mortality in individuals with diabetes mellitus. Endothelial injury is a major contributing factor for vascular dysfunction in diabetes. However, the precise mechanisms underlying endothelial cell injury and their heterogeneity in diabetes remains elusive. In this study, single-cell sequencing is performed in heart tissues from leptin receptor knock-out (db/db) diabetic mice at various pathological stages. Through cell cluster identification, differential gene analysis, intercellular communication analysis, pseudo time analysis, and transcription factor analysis, a novel mechanism of cardiac vascular endothelial damage in diabetes is identified. Specifically, a single-cell transcription map of cardiac vascular endothelial cells is presented in db/db mice. Diverse cellular clusters are found to play vital roles under diabetes-induced damage, highlighting crucial transcription factors involved in their regulation. In addition, the essential transcription factor Ets1 is found to protect against vascular endothelial injury in db/db mice. In summary, the work provides a comprehensive understanding of the development of diabetic cardiac vascular endothelial damage and the heterogeneity of the cells involved. These findings offer valuable insights into potential treatments and assessments of diabetic cardiovascular endothelial damage.
Keywords: cardiovascular; diabetic injury; endothelial cells; single‐cell analysis.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Sun H., Saeedi P., Karuranga S., Pinkepank M., Ogurtsova K., Duncan B. B., Stein C., Basit A., Chan J. C. N., Mbanya J. C., Pavkov M. E., Ramachandaran A., Wild S. H., James S., Herman W. H., Zhang P., Bommer C., Kuo S., Boyko E. J., Magliano D. J., Diabetes Res Clin Pract 2022, 183, 109119. - PMC - PubMed
-
- Syed F. Z., Ann. Intern. Med. 2022, 175, ITC33. - PubMed
MeSH terms
Substances
Grants and funding
- 82470358/National Natural Science Foundation of China
- 82070398/National Natural Science Foundation of China
- 81922008/National Natural Science Foundation of China
- 2022-JCJQ-ZD-095-00/Key basic research projects of basic strengthening plan
- BKJWS221J004/Key logistic research projects
- 2024RS-CXTD-78/Shaanxi Provincial Science and Technology Innovation Team
- Top Young Talents Special Support Program in Shaanxi Province
- XJZT24CZ13/Xijing Hospital Research Promotion Program
- XJZT24JC36/Xijing Hospital Research Promotion Program
- XJZT24LY38/Xijing Hospital Research Promotion Program
- LHJJ2023-YX05/Xijing Hospital Research Promotion Program
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
