Orthopteran Neo-Sex Chromosomes Reveal Dynamics of Recombination Suppression and Evolution of Supergenes
- PMID: 39475093
- PMCID: PMC11589690
- DOI: 10.1111/mec.17567
Orthopteran Neo-Sex Chromosomes Reveal Dynamics of Recombination Suppression and Evolution of Supergenes
Abstract
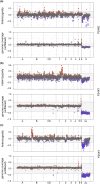
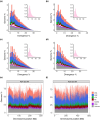
The early evolution of sex chromosomes has remained obscure for more than a century. The Vandiemenella viatica species group of morabine grasshoppers is highly suited for studying the early stages of sex chromosome divergence and degeneration of the Y chromosome. This stems from the fact that neo-XY sex chromosomes have independently evolved multiple times by X-autosome fusions with different autosomes. Here, we generated new chromosome-level assemblies for two chromosomal races representing karyotypes with and without neo-sex chromosomes (P24XY and P24X0), and sequence data of a third chromosomal race with a different neo-XY chromosome system (P25XY). Interestingly, these two neo-XY chromosomal races are formed by different X-autosome fusions (involving chr1 and chrB, respectively), and we found that both neo-Y chromosomes have partly ceased to recombine with their neo-X counterpart. We show that the neo-XY chromosomes have diverged through accumulation of SNPs and structural mutations, and that many neo-Y-linked genes have degenerated since recombination ceased. However, the non-recombining regions of neo-Y chromosomes host non-degenerated genes crucial for sex determination, such as sex-lethal and transformer, alongside genes associated with spermatogenesis, fertility, and reproduction, illustrating their integrative role as a masculinizing supergene. Contrary to expectations, the neo-Y chromosomes showed (slightly) lower density of transposable elements (TEs) compared to other genomic regions. The study reveals the unique dynamics of young sex chromosomes, with evolution of recombination suppression and pronounced decay of (some) neo-sex chromosome genes, and provides a compelling case illustrating how chromosomal fusions and post-fusion mutational processes contribute to the evolution of supergenes.
Keywords: chromosomal rearrangements; genetic degeneration; genomic recombination; neo‐sex chromosomes; repetitive DNA; sexual antagonistic locus; supergenes.
© 2024 The Author(s). Molecular Ecology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Molecular cytogenetic analysis reveals the existence of two independent neo-XY sex chromosome systems in Anatolian Pamphagidae grasshoppers.BMC Evol Biol. 2017 Feb 7;17(Suppl 1):20. doi: 10.1186/s12862-016-0868-9. BMC Evol Biol. 2017. PMID: 28251879 Free PMC article.
-
Uncovering the evolutionary history of neo-XY sex chromosomes in the grasshopper Ronderosia bergii (Orthoptera, Melanoplinae) through satellite DNA analysis.BMC Evol Biol. 2018 Jan 8;18(1):2. doi: 10.1186/s12862-017-1113-x. BMC Evol Biol. 2018. PMID: 29329524 Free PMC article.
-
Progressive recombination suppression and differentiation in recently evolved neo-sex chromosomes.Mol Biol Evol. 2013 May;30(5):1131-44. doi: 10.1093/molbev/mst035. Epub 2013 Feb 23. Mol Biol Evol. 2013. PMID: 23436913 Free PMC article.
-
[Role of transposons in origin and evolution of plant XY sex chromosomes].Yi Chuan. 2015 Feb;37(2):157-164. doi: 10.16288/j.yczz.14-305. Yi Chuan. 2015. PMID: 25665642 Review. Chinese.
-
The evolution of a neo-XY1Y2 sex chromosome system by autosome-sex chromosome fusion in Dundocoris nodulicarinus Jacobs (Heteroptera: Aradidae: Carventinae).Chromosome Res. 2004;12(2):175-91. doi: 10.1023/b:chro.0000013155.99614.57. Chromosome Res. 2004. PMID: 15053487 Review.
Cited by
-
Variable organization of repeats and hidden diversity of XY sex chromosomes in Pentatomidae true Bugs (Hemiptera) revealed through comparative genomic hybridization.Chromosoma. 2025 May 26;134(1):4. doi: 10.1007/s00412-025-00831-7. Chromosoma. 2025. PMID: 40418433
References
-
- Atchley, W. R. , and Cheney J.. 1974. “Morphometric Differentiation in the Viatica Group of Morabine Grasshoppers (Orthoptera, Eumastacidae).” Systematic Zoology 23, no. 3: 400–415. 10.2307/2412545. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

