Metabolic interplay between Proteus mirabilis and Enterococcus faecalis facilitates polymicrobial biofilm formation and invasive disease
- PMID: 39475232
- PMCID: PMC11640290
- DOI: 10.1128/mbio.02164-24
Metabolic interplay between Proteus mirabilis and Enterococcus faecalis facilitates polymicrobial biofilm formation and invasive disease
Abstract
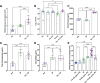
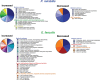
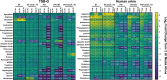
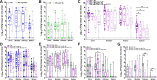
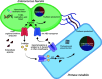
Biofilms play an important role in the development and pathogenesis of catheter-associated urinary tract infection (CAUTI). Proteus mirabilis and Enterococcus faecalis are common CAUTI pathogens that persistently co-colonize the catheterized urinary tract and form biofilms with increased biomass and antibiotic resistance. In this study, we uncover the metabolic interplay that drives biofilm enhancement and examine the contribution to CAUTI severity. Through compositional and proteomic biofilm analyses, we determined that the increase in biofilm biomass stems from an increase in the protein fraction of the polymicrobial biofilm. We further observed an enrichment in proteins associated with ornithine and arginine metabolism in polymicrobial biofilms compared with single-species biofilms. We show that arginine/ornithine antiport by E. faecalis promotes arginine biosynthesis and metabolism in P. mirabilis, ultimately driving the increase in polymicrobial biofilm protein content without affecting viability of either species. We further show that disrupting E. faecalis ornithine antiport alters the metabolic profile of polymicrobial biofilms and prevents enhancement, and this defect was complemented by supplementation with exogenous ornithine. In a murine model of CAUTI, ornithine antiport did not contribute to E. faecalis colonization but was required for the increased incidence of urinary stone formation and bacteremia that occurs during polymicrobial CAUTI with P. mirabilis. Thus, disrupting metabolic interplay between common co-colonizing species may represent a viable strategy for reducing risk of bacteremia.IMPORTANCEChronic infections often involve the formation of antibiotic-resistant biofilm communities that include multiple different microbes, which pose a challenge for effective treatment. In the catheterized urinary tract, potential pathogens persistently co-colonize for long periods of time and the interactions between them can lead to more severe disease outcomes. In this study, we identified the metabolite L-ornithine as a key mediator of disease-enhancing interactions between two common and challenging pathogens, Enterococcus faecalis and Proteus mirabilis. Disrupting ornithine-mediated interactions may therefore represent a strategy to prevent polymicrobial biofilm formation and decrease risk of severe disease.
Keywords: Enterococcus faecalis; Proteus mirabilis; arginine; bacteremia; bacterial metabolism; biofilm; catheter-associated urinary tract infection; ornithine; polymicrobial; urinary tract infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Update of
-
Metabolic interplay between Proteus mirabilis and Enterococcus faecalis facilitates polymicrobial biofilm formation and invasive disease.bioRxiv [Preprint]. 2024 Jun 4:2023.03.17.533237. doi: 10.1101/2023.03.17.533237. bioRxiv. 2024. Update in: mBio. 2024 Dec 11;15(12):e0216424. doi: 10.1128/mbio.02164-24. PMID: 36993593 Free PMC article. Updated. Preprint.
References
-
- Sabih A, Leslie SW. 2022. Complicated urinary tract infections. StatPearls Publishing, Treasure Island (FL). - PubMed
-
- Bono MJ, Leslie SW, Reygaert WC. 2022. Uncomplicated urinary tract infection. StatPearls Publishing, Treasure Island (FL). https://www.ncbi.nlm.nih.gov/books/NBK470195/. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
