Development of a suite of Proteus mirabilis-derived urea-inducible promoters
- PMID: 39475285
- PMCID: PMC11577795
- DOI: 10.1128/aem.01273-24
Development of a suite of Proteus mirabilis-derived urea-inducible promoters
Abstract
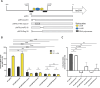
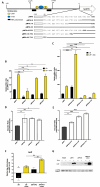
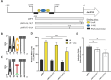
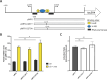
Catheter-associated urinary tract infections (CAUTIs) are a significant burden on healthcare systems, accounting for up to 40% of hospital-acquired infections globally. A prevalent CAUTI pathogen, Proteus mirabilis, is an understudied Gram-negative bacterium. One sequela of P. mirabilis CAUTI is the production of urinary stones, which complicates treatment and clearing of the infection. Stone formation is induced by the activity of urease, a nickel-metalloenzyme that is regulated by UreR in a urea-dependent manner. As urea is abundant in the urinary tract, urease genes are highly expressed during experimental UTI. We sought to leverage the urease promoter to create an expression system that would enable urea-inducible expression of genes during in vitro experiments as well as during experimental UTI. During preliminary studies, we observed unexpectedly high levels of basal expression of the urease promoter. This was somewhat dependent on the presence of regulator UreR. To further develop this expression system, we generated a series of reporter constructs to assess the impact of specific promoter elements on promoter activity in the presence and absence of urea. Elements of interest included known regulatory binding sites, alternative translational start sites, and single-nucleotide polymorphisms identified through comparative genomics. This work describes a suite of urea-inducible promoters, constructed during this study, that exhibit a variety of expression dynamics, providing a customizable platform for gene expression.IMPORTANCEUrea is an inexpensive molecule that can easily be supplied during in vitro experiments. A urea-inducible promoter would also be activated by environments where urea naturally occurs, such as in the urinary tract. Thus, the development of a urea-inducible system for selective gene expression is of great interest to the field of uropathogenesis as it would enable selective gene induction during experimental urinary tract infection. This expression system would also have important applications for recombinant protein production in biotech and manufacturing.
Keywords: Proteus mirabilis; inducible expression; promoter; urease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Davey LE, Malkus PN, Villa M, Dolat L, Holmes ZC, Letourneau J, Ansaldo E, David LA, Barton GM, Valdivia RH. 2023. A genetic system for Akkermansia muciniphila reveals a role for mucin foraging in gut colonization and host sterol biosynthesis gene expression. Nat Microbiol 8:1450–1467. doi:10.1038/s41564-023-01407-w - DOI - PMC - PubMed
-
- Maglennon GA, Cook BS, Deeney AS, Bossé JT, Peters SE, Langford PR, Maskell DJ, Tucker AW, Wren BW, Rycroft AN, BRaDP1T consortium . 2013. Transposon mutagenesis in Mycoplasma hyopneumoniae using a novel mariner-based system for generating random mutations. Vet Res 44:124. doi:10.1186/1297-9716-44-124 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

