Pangenome analysis of Paenibacillus polymyxa strains reveals the existence of multiple and functionally distinct Paenibacillus species
- PMID: 39475287
- PMCID: PMC11577871
- DOI: 10.1128/aem.01740-24
Pangenome analysis of Paenibacillus polymyxa strains reveals the existence of multiple and functionally distinct Paenibacillus species
Abstract
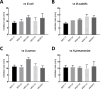
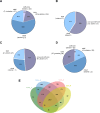
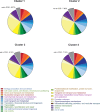
Paenibacillus polymyxa, a Gram-positive bacterium commonly found in soil and plant roots, plays an important role in the environment due to its nitrogen-fixing ability and is renowned for producing antibiotics like polymyxin. In this study, we present a robust framework for investigating the evolutionary and taxonomic connections of strains belonging to P. polymyxa available at the National Center for Biotechnology Information, as well as five new additional strains isolated at the University of Camerino (Italy), through pangenome analysis. These strains can produce secondary metabolites active against Staphylococcus aureus and Klebsiella pneumoniae. Employing techniques such as digital DNA-DNA hybridization (dDDH), average nucleotide identity (ANI) estimation, OrthoFinder, and ribosomal multilocus sequence typing, we consistently divided these P. polymyxa strains into four clusters, which differ significantly in terms of ANI and dDDH percentages, both considered as reference indices for separating bacterial species. Moreover, the strains of Cluster 2 were re-classified as belonging to the Paenibacillus ottowii species. By comparing the pangenomes, we identified the core genes of each cluster and analyzed them to recognize distinctive features in terms of biosynthetic/metabolic potential. The comparison of pangenomes also allowed us to pinpoint differences between clusters in terms of genetic variability and the percentage of the genome dedicated to core and accessory genes. In conclusion, the data obtained from our analyses of strains belonging to the P. polymyxa species converge toward a necessary reclassification, which will require a fundamental contribution from microbiologists in the near future.
Importance: The development of sequencing technologies has led to an exponential increase in microbial sequencing data. Accurately identifying bacterial species remains a challenge because of extensive intra-species variability, the need for multiple identification methods, and the rapid rate of taxonomic changes. A substantial contribution to elucidating the relationships among related bacterial strains comes from comparing their genomic sequences. This comparison also allows for the identification of the "pangenome," which is the set of genes shared by all individuals of a species, as well as the set of genes that are unique to subpopulations. Here, we applied this approach to Paenibacillus polymyxa, a species studied for its potential as a biofertilizer and biocontrol agent and known as an antibiotic producer. Our work highlights the need for a more efficient classification of this bacterial species and provides a better delineation of strains with different properties.
Keywords: Paenibacillus polymyxa; evolutionary analysis; genomics; pangenome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

