NUP98 fusion proteins and KMT2A-MENIN antagonize PRC1.1 to drive gene expression in AML
- PMID: 39475509
- PMCID: PMC11780541
- DOI: 10.1016/j.celrep.2024.114901
NUP98 fusion proteins and KMT2A-MENIN antagonize PRC1.1 to drive gene expression in AML
Abstract
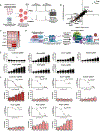
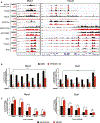
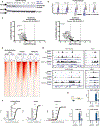
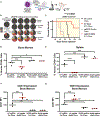
Control of stem cell-associated genes by Trithorax group (TrxG) and Polycomb group (PcG) proteins is frequently misregulated in cancer. In leukemia, oncogenic fusion proteins hijack the TrxG homolog KMT2A and disrupt PcG activity to maintain pro-leukemogenic gene expression, though the mechanisms by which oncofusion proteins antagonize PcG proteins remain unclear. Here, we define the relationship between NUP98 oncofusion proteins and the non-canonical polycomb repressive complex 1.1 (PRC1.1) in leukemia using Menin-KMT2A inhibitors and targeted degradation of NUP98 fusion proteins. Eviction of the NUP98 fusion-Menin-KMT2A complex from chromatin is not sufficient to silence pro-leukemogenic genes. In the absence of PRC1.1, key oncogenes remain transcriptionally active. Transition to a repressed chromatin state requires the accumulation of PRC1.1 and repressive histone modifications. We show that PRC1.1 loss leads to resistance to small-molecule Menin-KMT2A inhibitors in vivo. Therefore, a critical function of oncofusion proteins that hijack Menin-KMT2A activity is antagonizing repressive chromatin complexes.
Keywords: CP: Cancer; CP: Molecular biology; acute myeloid leukemia; chromatin complex; lysine methyltransferase 2A; menin; nascent transcriptomics; non-canonical polycomb repressive complex 1.1; nucleoporin 98-rearrangement; oncogenic fusion protein.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.A.A. has been a consultant and/or shareholder for Neomorph, Imago Biosciences, Hyku Therapeutics, C4 Therapeutics, Accent Therapeutics, and Nimbus Therapeutics. S.A.A. has received research support from Janssen and Syndax. S.A.A. is an inventor on a patent related to MENIN inhibition WO/2017/132398A1.
Figures



References
-
- Lewis EB (1978). A gene complex controlling segmentation in Drosophila. Nature 276, 565–570. - PubMed
-
- Struhl G (1981). A gene product required for correct initiation of segmental determination in Drosophila. Nature 293, 36–41. - PubMed
-
- Ingham PW (1983). Differential expression of bithorax complex genes in the absence of the extra sex combs and trithorax genes. Nature 306, 591–593. - PubMed
-
- Schuettengruber B, Bourbon HM, Di Croce L, and Cavalli G (2017). Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 171, 34–57. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

