Caspase-2 kills cells with extra centrosomes
- PMID: 39475598
- PMCID: PMC11524169
- DOI: 10.1126/sciadv.ado6607
Caspase-2 kills cells with extra centrosomes
Abstract
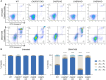
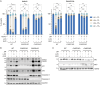
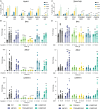
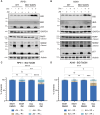
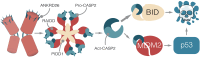
Centrosomes are membrane-less organelles that orchestrate a wide array of biological functions by acting as microtubule organizing centers. Here, we report that caspase-2-driven apoptosis is elicited in blood cells failing cytokinesis and that extra centrosomes are necessary to trigger this cell death. Activation of caspase-2 depends on the PIDDosome multi-protein complex, and priming of PIDD1 at extra centrosomes is necessary for pathway activation. Accordingly, loss of its centrosomal adapter, ANKRD26, allows for cell survival and unrestricted polyploidization in response to cytokinesis failure. Mechanistically, cell death is initiated upstream of mitochondria via caspase-2-mediated processing of the BCL2 family protein BID, driving BAX/BAK-dependent mitochondrial outer membrane permeabilization (MOMP). Remarkably, BID-deficient cells enforce apoptosis by engaging p53-dependent proapoptotic transcriptional responses initiated by caspase-2. Consistently, BID and MDM2 act as shared caspase-2 substrates, with BID being kinetically favored. Our findings document that the centrosome limits its own unscheduled duplication by the induction of PIDDosome-driven mitochondrial apoptosis to avoid potentially pathogenic polyploidization events.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

