Multi-Center Fetal Brain Tissue Annotation (FeTA) Challenge 2022 Results
- PMID: 39475746
- PMCID: PMC12099263
- DOI: 10.1109/TMI.2024.3485554
Multi-Center Fetal Brain Tissue Annotation (FeTA) Challenge 2022 Results
Abstract
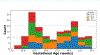
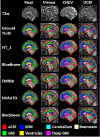
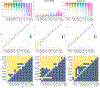
Segmentation is a critical step in analyzing the developing human fetal brain. There have been vast improvements in automatic segmentation methods in the past several years, and the Fetal Brain Tissue Annotation (FeTA) Challenge 2021 helped to establish an excellent standard of fetal brain segmentation. However, FeTA 2021 was a single center study, limiting real-world clinical applicability and acceptance. The multi-center FeTA Challenge 2022 focused on advancing the generalizability of fetal brain segmentation algorithms for magnetic resonance imaging (MRI). In FeTA 2022, the training dataset contained images and corresponding manually annotated multi-class labels from two imaging centers, and the testing data contained images from these two centers as well as two additional unseen centers. The multi-center data included different MR scanners, imaging parameters, and fetal brain super-resolution algorithms applied. 16 teams participated and 17 algorithms were evaluated. Here, the challenge results are presented, focusing on the generalizability of the submissions. Both in- and out-of-domain, the white matter and ventricles were segmented with the highest accuracy (Top Dice scores: 0.89, 0.87 respectively), while the most challenging structure remains the grey matter (Top Dice score: 0.75) due to anatomical complexity. The top 5 average Dices scores ranged from 0.81-0.82, the top 5 average percentile Hausdorff distance values ranged from 2.3-2.5mm, and the top 5 volumetric similarity scores ranged from 0.90-0.92. The FeTA Challenge 2022 was able to successfully evaluate and advance generalizability of multi-class fetal brain tissue segmentation algorithms for MRI and it continues to benchmark new algorithms.
Figures


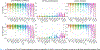

References
-
- Glocker B, Robinson R, Castro DC, Dou Q, and Konukoglu E, “Machine learning with multi-site imaging data: An empirical study on the impact of scanner effects,” 2019, arXiv:1910.04597.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

