Spatial transcriptomics of meningeal inflammation reveals inflammatory gene signatures in adjacent brain parenchyma
- PMID: 39475792
- PMCID: PMC11524578
- DOI: 10.7554/eLife.88414
Spatial transcriptomics of meningeal inflammation reveals inflammatory gene signatures in adjacent brain parenchyma
Abstract
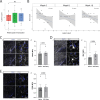
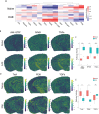
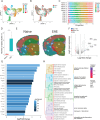
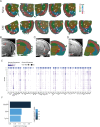
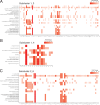
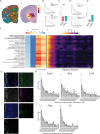
While modern high efficacy disease modifying therapies have revolutionized the treatment of relapsing-remitting multiple sclerosis, they are less effective at controlling progressive forms of the disease. Meningeal inflammation is a recognized risk factor for cortical gray matter pathology which can result in disabling symptoms such as cognitive impairment and depression, but the mechanisms linking meningeal inflammation and gray matter pathology remain unclear. Here, we performed magnetic resonance imaging (MRI)-guided spatial transcriptomics in a mouse model of autoimmune meningeal inflammation to characterize the transcriptional signature in areas of meningeal inflammation and the underlying brain parenchyma. We found broadly increased activity of inflammatory signaling pathways at sites of meningeal inflammation, but only a subset of these pathways active in the adjacent brain parenchyma. Subclustering of regions adjacent to meningeal inflammation revealed the subset of immune programs induced in brain parenchyma, notably complement signaling and antigen processing/presentation. Trajectory gene and gene set modeling analysis confirmed variable penetration of immune signatures originating from meningeal inflammation into the adjacent brain tissue. This work contributes a valuable data resource to the field, provides the first detailed spatial transcriptomic characterization in a model of meningeal inflammation, and highlights several candidate pathways in the pathogenesis of gray matter pathology.
Keywords: experimental autoimmune encephalomyelitis; gray matter degeneration; immunology; inflammation; meningeal inflammation; mouse; multiple sclerosis; neuroscience; spatial transcriptomics.
© 2023, Gadani, Singh et al.
Conflict of interest statement
SG, SS, SK, JH, MS, PC, PB No competing interests declared
Figures





Update of
- doi: 10.1101/2023.06.02.543421
- doi: 10.7554/eLife.88414.1
- doi: 10.7554/eLife.88414.2
- doi: 10.7554/eLife.88414.3
References
-
- Absinta M, Vuolo L, Rao A, Nair G, Sati P, Cortese ICM, Ohayon J, Fenton K, Reyes-Mantilla MI, Maric D, Calabresi PA, Butman JA, Pardo CA, Reich DS. Gadolinium-based MRI characterization of leptomeningeal inflammation in multiple sclerosis. Neurology. 2015;85:18–28. doi: 10.1212/WNL.0000000000001587. - DOI - PMC - PubMed
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. - DOI - PMC - PubMed
-
- Bedussi B, van der Wel NN, de Vos J, van Veen H, Siebes M, VanBavel E, Bakker EN. Paravascular channels, cisterns, and the subarachnoid space in the rat brain: a single compartment with preferential pathways. Journal of Cerebral Blood Flow and Metabolism. 2017;37:1374–1385. doi: 10.1177/0271678X16655550. - DOI - PMC - PubMed
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

