PilY1 regulates the dynamic architecture of the type IV pilus machine in Pseudomonas aeruginosa
- PMID: 39477930
- PMCID: PMC11525922
- DOI: 10.1038/s41467-024-53638-y
PilY1 regulates the dynamic architecture of the type IV pilus machine in Pseudomonas aeruginosa
Abstract
Type IV pili (T4P) produced by the pathogen Pseudomonas aeruginosa play a pivotal role in adhesion, surface motility, biofilm formation, and infection in humans. Despite the significance of T4P as a potential therapeutic target, key details of their dynamic assembly and underlying molecular mechanisms of pilus extension and retraction remain elusive, primarily due to challenges in isolating intact T4P machines from the bacterial cell envelope. Here, we combine cryo-electron tomography with subtomogram averaging and integrative modelling to resolve in-situ architectural details of the dynamic T4P machine in P. aeruginosa cells. The T4P machine forms 7-fold symmetric cage-like structures anchored in the cell envelope, providing a molecular framework for the rapid exchange of major pilin subunits during pilus extension and retraction. Our data suggest that the T4P adhesin PilY1 forms a champagne-cork-shaped structure, effectively blocking the secretin channel in the outer membrane whereas the minor-pilin complex in the periplasm appears to contact PilY1 via the central pore of the secretin gate. These findings point to a hypothetical model where the interplay between the secretin protein PilQ and the PilY1-minor-pilin priming complex is important for optimizing conformations of the T4P machine in P. aeruginosa, suggesting a gate-keeping mechanism that regulates pilus dynamics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


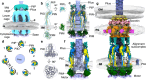


References
-
- Craig, L., Pique, M. E. & Tainer, J. A. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol.2, 363–378 (2004). - PubMed
-
- Burrows, L. L. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol.66, 493–520 (2012). - PubMed
-
- Ellison, C. K., Whitfield, G. B. & Brun, Y. V. Type IV pili: dynamic bacterial nanomachines. FEMS Microbiol. Rev.46, 10.1093/femsre/fuab053 (2022). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

