Molecular mimicry as a mechanism of viral immune evasion and autoimmunity
- PMID: 39477943
- PMCID: PMC11526117
- DOI: 10.1038/s41467-024-53658-8
Molecular mimicry as a mechanism of viral immune evasion and autoimmunity
Abstract
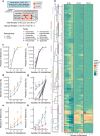
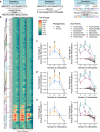
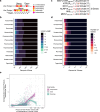
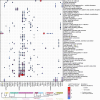
Mimicry of host protein structures, or 'molecular mimicry', is a common mechanism employed by viruses to evade the host's immune system. Short linear amino acid (AA) molecular mimics can elicit cross-reactive antibodies and T cells from the host, but the prevalence of such mimics throughout the human virome has not been fully explored. Here we evaluate 134 human-infecting viruses and find significant usage of linear mimicry across the virome, particularly those in the Herpesviridae and Poxviridae families. Furthermore, host proteins related to cellular replication and inflammation, autosomes, the X chromosome, and thymic cells are enriched as viral mimicry targets. Finally, we find that short linear mimicry from Epstein-Barr virus (EBV) is higher in auto-antibodies found in patients with multiple sclerosis than previously appreciated. Our results thus hint that human-infecting viruses leverage mimicry in the course of their infection, and that such mimicry may contribute to autoimmunity, thereby prompting potential targets for therapies.
© 2024. The Author(s).
Conflict of interest statement
E.M. has received research funding from Babson Diagnostics, honorarium from Multiple Sclerosis Association of America and has served on advisory boards of Genentech, Horizon, Teva and Viela Bio. The other authors declare no competing interests.
Figures

Update of
-
Molecular Mimicry as a Mechanism of Viral Immune Evasion and Autoimmunity.bioRxiv [Preprint]. 2024 Mar 9:2024.03.08.583134. doi: 10.1101/2024.03.08.583134. bioRxiv. 2024. Update in: Nat Commun. 2024 Oct 30;15(1):9403. doi: 10.1038/s41467-024-53658-8. PMID: 38496443 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
- 5T32DA018926-18/U.S. Department of Health & Human Services | NIH | National Institute on Drug Abuse (NIDA)
- K08 T26-1616-11/U.S. Department of Health & Human Services | NIH | National Institute on Alcohol Abuse and Alcoholism (NIAAA)
- R01 AI104870/AI/NIAID NIH HHS/United States
- T32 DA018926/DA/NIDA NIH HHS/United States
- R01AI104870-S1/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources

