Single-cell landscape of innate and acquired drug resistance in acute myeloid leukemia
- PMID: 39477946
- PMCID: PMC11525670
- DOI: 10.1038/s41467-024-53535-4
Single-cell landscape of innate and acquired drug resistance in acute myeloid leukemia
Abstract
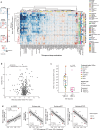
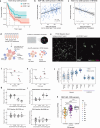
Deep single-cell multi-omic profiling offers a promising approach to understand and overcome drug resistance in relapsed or refractory (rr) acute myeloid leukemia (AML). Here, we combine single-cell ex vivo drug profiling (pharmacoscopy) with single-cell and bulk DNA, RNA, and protein analyses, alongside clinical data from 21 rrAML patients. Unsupervised data integration reveals reduced ex vivo response to the Bcl-2 inhibitor venetoclax (VEN) in patients treated with both a hypomethylating agent (HMA) and VEN, compared to those pre-exposed to chemotherapy or HMA alone. Integrative analysis identifies both known and unreported mechanisms of innate and treatment-related VEN resistance and suggests alternative treatments, like targeting increased proliferation with the PLK inhibitor volasertib. Additionally, high CD36 expression in VEN-resistant blasts associates with sensitivity to CD36-targeted antibody treatment ex vivo. This study demonstrates how single-cell multi-omic profiling can uncover drug resistance mechanisms and treatment vulnerabilities, providing a valuable resource for future AML research.
© 2024. The Author(s).
Conflict of interest statement
B.B. has founded and is a shareholder and member of the board of Navignostics, a precision oncology spin-off from the University of Zurich. B.B. reports personal consultancy fees as Advisory board member from Standard BioTools. B.S. was a scientific co-founder of Allcyte, which has been acquired by Exscientia. B.S. is a shareholder of Exscientia and a co-inventor on US patent application 15/514,045 relevant to the study. L.P. and G.G. are listed as inventors on patents related to the 4i technology (WO 2019/207004; WO 2020/008071). G.R. is listed on a patent application related to single-cell analyses (European Patent Application No. 20170724.7), and G.R. is cofounder and on the Scientific Advisory Board of Computomics GmbH. H.M. is on advisory boards for Bayer, Astra Zeneca, Janssen, Roche, and Merck. M.P.L. is scientific advisor and board member of Oncobit and has received unrelated research funding from Roche, Novartis, Scailyte, Molecular Partners, and Bacoba. V.H.K. is an invited speaker for Sharing Progress in Cancer Care (SPCC) and Indica Labs; advisory board of Takeda; sponsored research agreements with Roche and IAG all unrelated to the current study; listed as innovator on patent applications in computational pathology. The Tumor Profiler study is jointly funded by a public-private partnership involving F. Hoffmann-La Roche Ltd., ETH Zurich, University of Zurich, University Hospital Zurich, and University Hospital Basel. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

