CCL3 predicts exceptional response to TGFβ inhibition in basal-like pancreatic cancer enriched in LIF-producing macrophages
- PMID: 39478186
- PMCID: PMC11525688
- DOI: 10.1038/s41698-024-00742-3
CCL3 predicts exceptional response to TGFβ inhibition in basal-like pancreatic cancer enriched in LIF-producing macrophages
Abstract
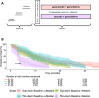
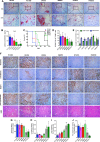
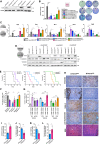
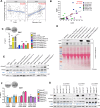
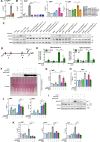
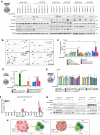
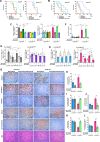
The TGFβ receptor inhibitor galunisertib showed promising efficacy in patients with pancreatic ductal adenocarcinoma (PDAC) in the phase 2 H9H-MC-JBAJ study. Identifying biomarkers for this treatment remains essential. Baseline plasma levels of chemokine CCL3 were integrated with clinical outcomes in PDAC patients treated with galunisertib plus gemcitabine (n = 104) or placebo plus gemcitabine (n = 52). High CCL3 was a poor prognostic factor in the placebo group (mOS 3.6 vs. 10.1 months; p < 0.01) but a positive predictor for galunisertib (mOS 9.2 vs. 3.6 months; p < 0.01). Mechanistically, tumor-derived CCL3 activates Tgfβ signaling in macrophages, inducing their M2 phenotype and Lif secretion, sustaining a mesenchymal/basal-like ecotype. TGFβ inhibition redirects macrophage polarization to M1, reducing Lif and shifting PDAC cells to a more epithelial/classical phenotype, improving gemcitabine sensitivity. This study supports exploring TGFβ-targeting agents in PDAC with a mesenchymal/basal-like ecotype driven by high CCL3 levels.
© 2024. The Author(s).
Conflict of interest statement
DM received honoraria as an advisory board member or consultant from Servier, Incyte, Tahio, iOnctura, Eli Lilly, Evotec, Shire, Baxter; received institutional support for research project from Shire, Celgene, Incyte, iOnctura, Roche. The other authors have declared that no conflict of interest exists.
Figures
References
-
- The Global Cancer Observatory. Pancreas, https://gco.iarc.fr/today/data/factsheets/cancers/13-Pancreas-fact-sheet... (2020).
-
- Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin.74, 12–49 (2024). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

