GASIDN: identification of sub-Golgi proteins with multi-scale feature fusion
- PMID: 39478465
- PMCID: PMC11526662
- DOI: 10.1186/s12864-024-10954-3
GASIDN: identification of sub-Golgi proteins with multi-scale feature fusion
Abstract
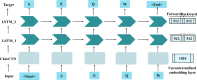
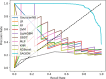
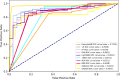
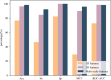
The Golgi apparatus is a crucial component of the inner membrane system in eukaryotic cells, playing a central role in protein biosynthesis. Dysfunction of the Golgi apparatus has been linked to neurodegenerative diseases. Accurate identification of sub-Golgi protein types is therefore essential for developing effective treatments for such diseases. Due to the expensive and time-consuming nature of experimental methods for identifying sub-Golgi protein types, various computational methods have been developed as identification tools. However, the majority of these methods rely solely on neighboring features in the protein sequence and neglect the crucial spatial structure information of the protein.To discover alternative methods for accurately identifying sub-Golgi proteins, we have developed a model called GASIDN. The GASIDN model extracts multi-dimension features by utilizing a 1D convolution module on protein sequences and a graph learning module on contact maps constructed from AlphaFold2.The model utilizes the deep representation learning model SeqVec to initialize protein sequences. GASIDN achieved accuracy values of 98.4% and 96.4% in independent testing and ten-fold cross-validation, respectively, outperforming the majority of previous predictors. To the best of our knowledge, this is the first method that utilizes multi-scale feature fusion to identify and locate sub-Golgi proteins. In order to assess the generalizability and scalability of our model, we conducted experiments to apply it in the identification of proteins from other organelles, including plant vacuoles and peroxisomes. The results obtained from these experiments demonstrated promising outcomes, indicating the effectiveness and versatility of our model. The source code and datasets can be accessed at https://github.com/SJNNNN/GASIDN .
Keywords: AlphaFold2; Deep representation learning; Graph neural network; Multi-scale features; Sub-Golgi proteins; TextCNN.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Yang W, Zhu X-J, Huang J, Ding H, Lin H. A brief survey of machine learning methods in protein sub-Golgi localization. Curr Bioinform. 2019;14(3):234–40. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

